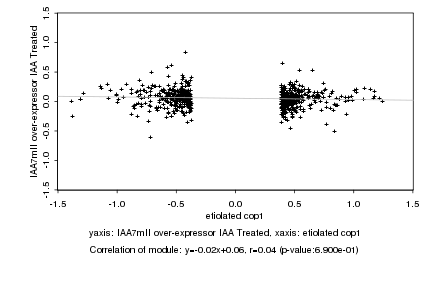
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
etiolated cop1 |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
1.238 |
0.005 |
| 2 |
245703_at |
AT5G04380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
1.211 |
0.063 |
| 3 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.170 |
0.176 |
| 4 |
265611_at |
AT2G25510
|
unknown |
1.169 |
0.096 |
| 5 |
266293_at |
AT2G29360
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.161 |
0.055 |
| 6 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
1.137 |
0.219 |
| 7 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
1.087 |
0.230 |
| 8 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
1.078 |
0.050 |
| 9 |
252983_at |
AT4G37980
|
ATCAD7, CAD7, CINNAMYL-ALCOHOL DEHYDROGENASE 7, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-1, elicitor-activated gene 3-1 |
1.021 |
0.211 |
| 10 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
1.019 |
0.159 |
| 11 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
1.002 |
0.197 |
| 12 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
0.987 |
0.029 |
| 13 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
0.973 |
0.086 |
| 14 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
0.955 |
0.070 |
| 15 |
255852_at |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
0.948 |
0.020 |
| 16 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
0.932 |
-0.219 |
| 17 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.930 |
0.013 |
| 18 |
245460_at |
AT4G16990
|
RLM3, RESISTANCE TO LEPTOSPHAERIA MACULANS 3 |
0.929 |
0.079 |
| 19 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.895 |
0.102 |
| 20 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.879 |
0.047 |
| 21 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.858 |
-0.054 |
| 22 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.849 |
0.065 |
| 23 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.846 |
-0.061 |
| 24 |
251309_at |
AT3G61220
|
SDR1, short-chain dehydrogenase/reductase 1 |
0.838 |
-0.051 |
| 25 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.833 |
-0.508 |
| 26 |
249355_at |
AT5G40500
|
unknown |
0.830 |
-0.035 |
| 27 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.824 |
-0.146 |
| 28 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
0.818 |
0.177 |
| 29 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
0.804 |
0.143 |
| 30 |
251658_at |
AT3G57020
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.797 |
-0.108 |
| 31 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.789 |
0.087 |
| 32 |
249128_at |
AT5G43440
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.767 |
-0.380 |
| 33 |
247218_at |
AT5G65010
|
ASN2, asparagine synthetase 2 |
0.764 |
-0.010 |
| 34 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
0.764 |
-0.090 |
| 35 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.755 |
0.214 |
| 36 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
0.747 |
0.198 |
| 37 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
0.743 |
0.081 |
| 38 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
0.737 |
0.079 |
| 39 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
0.737 |
0.310 |
| 40 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
0.735 |
0.143 |
| 41 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
0.723 |
0.157 |
| 42 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.720 |
-0.136 |
| 43 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.712 |
0.009 |
| 44 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.704 |
0.101 |
| 45 |
250135_at |
AT5G15360
|
unknown |
0.702 |
0.118 |
| 46 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
0.698 |
0.059 |
| 47 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
0.693 |
0.125 |
| 48 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
0.690 |
0.164 |
| 49 |
266572_at |
AT2G23840
|
[HNH endonuclease] |
0.687 |
0.079 |
| 50 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.684 |
0.079 |
| 51 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
0.679 |
0.098 |
| 52 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.670 |
-0.009 |
| 53 |
262642_at |
AT1G62690
|
unknown |
0.665 |
0.031 |
| 54 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
0.663 |
0.060 |
| 55 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
0.662 |
0.158 |
| 56 |
252010_at |
AT3G52740
|
unknown |
0.653 |
0.117 |
| 57 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
0.648 |
0.541 |
| 58 |
248873_at |
AT5G46450
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.648 |
0.099 |
| 59 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.635 |
-0.034 |
| 60 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.630 |
-0.111 |
| 61 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
0.626 |
-0.057 |
| 62 |
261751_at |
AT1G76080
|
ATCDSP32, ARABIDOPSIS THALIANA CHLOROPLASTIC DROUGHT-INDUCED STRESS PROTEIN OF 32 KD, CDSP32, chloroplastic drought-induced stress protein of 32 kD |
0.625 |
0.183 |
| 63 |
246792_at |
AT5G27290
|
unknown |
0.622 |
0.101 |
| 64 |
264636_at |
AT1G65490
|
unknown |
0.616 |
0.160 |
| 65 |
252366_at |
AT3G48420
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.616 |
0.209 |
| 66 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
0.614 |
0.156 |
| 67 |
267057_at |
AT2G32500
|
[Stress responsive alpha-beta barrel domain protein] |
0.612 |
0.073 |
| 68 |
254251_at |
AT4G23300
|
CRK22, cysteine-rich RLK (RECEPTOR-like protein kinase) 22 |
0.611 |
0.221 |
| 69 |
248709_at |
AT5G48470
|
PRDA1, PEP-Related Development Arrested 1 |
0.611 |
0.061 |
| 70 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.606 |
0.117 |
| 71 |
251935_at |
AT3G54090
|
FLN1, fructokinase-like 1 |
0.586 |
0.129 |
| 72 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.582 |
0.217 |
| 73 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.579 |
0.124 |
| 74 |
264357_at |
AT1G03360
|
ATRRP4, ribosomal RNA processing 4, RRP4, ribosomal RNA processing 4 |
0.575 |
-0.162 |
| 75 |
246268_at |
AT1G31800
|
CYP97A3, cytochrome P450, family 97, subfamily A, polypeptide 3, LUT5, LUTEIN DEFICIENT 5 |
0.571 |
-0.127 |
| 76 |
244994_at |
ATCG01010
|
NDHF |
0.570 |
0.026 |
| 77 |
250762_at |
AT5G05990
|
[Mitochondrial glycoprotein family protein] |
0.566 |
0.081 |
| 78 |
267505_at |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
0.564 |
0.259 |
| 79 |
253391_at |
AT4G32590
|
[2Fe-2S ferredoxin-like superfamily protein] |
0.562 |
0.184 |
| 80 |
245029_at |
AT2G26580
|
YAB5, YABBY5 |
0.562 |
0.042 |
| 81 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
0.556 |
-0.008 |
| 82 |
245417_at |
AT4G17360
|
[Formyl transferase] |
0.556 |
0.178 |
| 83 |
248105_at |
AT5G55280
|
ATFTSZ1-1, ARABIDOPSIS THALIANA HOMOLOG OF BACTERIAL CYTOKINESIS Z-RING PROTEIN FTSZ 1-1, CPFTSZ, CHLOROPLAST FTSZ, FTSZ1-1, homolog of bacterial cytokinesis Z-ring protein FTSZ 1-1 |
0.553 |
0.090 |
| 84 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.553 |
0.118 |
| 85 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
0.551 |
0.195 |
| 86 |
258607_at |
AT3G02730
|
ATF1, TRXF1, thioredoxin F-type 1 |
0.548 |
-0.080 |
| 87 |
247491_at |
AT5G61880
|
[Protein Transporter, Pam16] |
0.544 |
-0.121 |
| 88 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
0.542 |
-0.257 |
| 89 |
261898_at |
AT1G80720
|
[Mitochondrial glycoprotein family protein] |
0.541 |
-0.009 |
| 90 |
261068_at |
AT1G07450
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.540 |
0.052 |
| 91 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
0.540 |
0.161 |
| 92 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.539 |
0.119 |
| 93 |
262745_at |
AT1G28600
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.538 |
0.533 |
| 94 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.538 |
0.286 |
| 95 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
0.535 |
0.044 |
| 96 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.532 |
0.062 |
| 97 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.531 |
0.212 |
| 98 |
248201_at |
AT5G54180
|
PTAC15, plastid transcriptionally active 15 |
0.527 |
0.091 |
| 99 |
246411_at |
AT1G57770
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.527 |
0.006 |
| 100 |
249554_at |
AT5G38290
|
[Peptidyl-tRNA hydrolase family protein] |
0.526 |
0.165 |
| 101 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
0.525 |
0.189 |
| 102 |
262693_at |
AT1G62780
|
unknown |
0.525 |
0.171 |
| 103 |
259444_at |
AT1G02370
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.525 |
0.062 |
| 104 |
261218_at |
AT1G20020
|
ATLFNR2, LEAF FNR 2, FNR2, ferredoxin-NADP(+)-oxidoreductase 2, LFNR2, leaf-type chloroplast-targeted FNR 2 |
0.524 |
0.048 |
| 105 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
0.523 |
-0.091 |
| 106 |
251820_at |
AT3G55040
|
GSTL2, glutathione transferase lambda 2 |
0.521 |
0.089 |
| 107 |
249404_at |
AT5G40160
|
EMB139, EMBRYO DEFECTIVE 139, EMB506, embryo defective 506 |
0.514 |
0.069 |
| 108 |
250327_at |
AT5G12050
|
unknown |
0.514 |
0.008 |
| 109 |
251142_at |
AT5G01015
|
unknown |
0.512 |
0.102 |
| 110 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
0.512 |
-0.186 |
| 111 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
0.510 |
0.346 |
| 112 |
245426_at |
AT4G17540
|
unknown |
0.510 |
-0.088 |
| 113 |
260688_at |
AT1G17665
|
unknown |
0.509 |
-0.029 |
| 114 |
254737_at |
AT4G13840
|
CER26, ECERIFERUM 26 |
0.508 |
0.126 |
| 115 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
0.507 |
0.070 |
| 116 |
259363_at |
AT1G13270
|
MAP1B, METHIONINE AMINOPEPTIDASE 1B, MAP1C, methionine aminopeptidase 1B |
0.507 |
-0.148 |
| 117 |
247610_at |
AT5G60630
|
unknown |
0.506 |
0.134 |
| 118 |
257131_at |
AT3G20240
|
[Mitochondrial substrate carrier family protein] |
0.506 |
0.188 |
| 119 |
262629_at |
AT1G06460
|
ACD31.2, ALPHA-CRYSTALLIN DOMAIN 31.2, ACD32.1, alpha-crystallin domain 32.1 |
0.504 |
-0.164 |
| 120 |
258884_at |
AT3G10050
|
OMR1, L-O-methylthreonine resistant 1 |
0.504 |
0.088 |
| 121 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
0.502 |
0.230 |
| 122 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
0.501 |
0.004 |
| 123 |
251727_at |
AT3G56290
|
unknown |
0.498 |
0.065 |
| 124 |
258929_at |
AT3G10060
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.497 |
-0.003 |
| 125 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.497 |
0.244 |
| 126 |
258223_at |
AT3G15840
|
PIFI, post-illumination chlorophyll fluorescence increase |
0.497 |
0.200 |
| 127 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.496 |
0.303 |
| 128 |
250087_at |
AT5G17270
|
[Protein prenylyltransferase superfamily protein] |
0.496 |
0.100 |
| 129 |
266015_at |
AT2G24190
|
SDR2, short-chain dehydrogenase/reductase 2 |
0.495 |
0.200 |
| 130 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
0.494 |
0.082 |
| 131 |
266264_at |
AT2G27775
|
unknown |
0.492 |
0.071 |
| 132 |
258380_at |
AT3G16650
|
PRL2, Pleiotropic regulatory locus 2 |
0.491 |
-0.127 |
| 133 |
258424_at |
AT3G16750
|
unknown |
0.490 |
-0.119 |
| 134 |
261296_at |
AT1G48460
|
unknown |
0.489 |
-0.041 |
| 135 |
254460_at |
AT4G21210
|
ATRP1, PPDK regulatory protein, RP1, PPDK regulatory protein |
0.487 |
0.104 |
| 136 |
260036_at |
AT1G68830
|
STN7, STT7 homolog STN7 |
0.486 |
0.032 |
| 137 |
253337_at |
AT4G33470
|
ATHDA14, HDA14, histone deacetylase 14 |
0.486 |
-0.040 |
| 138 |
260696_at |
AT1G32520
|
unknown |
0.485 |
0.010 |
| 139 |
265819_at |
AT2G17972
|
unknown |
0.485 |
-0.115 |
| 140 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
0.484 |
0.010 |
| 141 |
256097_at |
AT1G13670
|
unknown |
0.481 |
0.235 |
| 142 |
246094_at |
AT5G19300
|
unknown |
0.481 |
-0.264 |
| 143 |
248854_at |
AT5G46580
|
[pentatricopeptide (PPR) repeat-containing protein] |
0.479 |
-0.099 |
| 144 |
257724_at |
AT3G18510
|
unknown |
0.479 |
-0.031 |
| 145 |
247709_at |
AT5G59250
|
[Major facilitator superfamily protein] |
0.479 |
-0.124 |
| 146 |
261638_at |
AT1G49975
|
[INVOLVED IN: photosynthesis] |
0.478 |
0.261 |
| 147 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
0.478 |
0.300 |
| 148 |
263533_at |
AT2G24820
|
AtTic55, translocon at the inner envelope membrane of chloroplasts 55, Tic55, translocon at the inner envelope membrane of chloroplasts 55, TIC55-II, translocon at the inner envelope membrane of chloroplasts 55-II |
0.477 |
-0.213 |
| 149 |
257647_at |
AT3G25805
|
unknown |
0.477 |
-0.242 |
| 150 |
248128_at |
AT5G54770
|
THI1, THI4, THIAMINE4, TZ, THIAZOLE REQUIRING |
0.477 |
0.275 |
| 151 |
267005_at |
AT2G34460
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.476 |
-0.013 |
| 152 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.476 |
0.038 |
| 153 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
0.476 |
0.150 |
| 154 |
252468_at |
AT3G46970
|
ATPHS2, Arabidopsis thaliana alpha-glucan phosphorylase 2, PHS2, alpha-glucan phosphorylase 2 |
0.475 |
-0.016 |
| 155 |
266265_at |
AT2G29340
|
[NAD-dependent epimerase/dehydratase family protein] |
0.475 |
0.297 |
| 156 |
257149_at |
AT3G27280
|
ATPHB4, prohibitin 4, PHB4, prohibitin 4 |
0.475 |
0.088 |
| 157 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
0.474 |
-0.030 |
| 158 |
263442_at |
AT2G28605
|
[Photosystem II reaction center PsbP family protein] |
0.472 |
0.044 |
| 159 |
266801_at |
AT2G22870
|
EMB2001, embryo defective 2001 |
0.471 |
-0.053 |
| 160 |
253782_at |
AT4G28590
|
AtECB1, ECB1, early chloroplast biogenesis 1, MRL7, Mesophyll-cell RNAi Library line 7, SVR4, SUPPRESSOR OF VARIEGATION 4 |
0.470 |
0.110 |
| 161 |
248537_at |
AT5G50100
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.470 |
-0.050 |
| 162 |
252410_at |
AT3G47450
|
ATNOA1, ATNOS1, NOA1, NO ASSOCIATED 1, NOS1, NITRIC OXIDE SYNTHASE 1, RIF1, RESISTANT TO INHIBITION WITH FOSMIDOMYCIN 1 |
0.469 |
-0.020 |
| 163 |
265869_at |
AT2G01760
|
ARR14, response regulator 14, RR14, response regulator 14 |
0.467 |
0.168 |
| 164 |
246651_at |
AT5G35170
|
[adenylate kinase family protein] |
0.466 |
-0.174 |
| 165 |
245909_at |
AT5G09380
|
[RNA polymerase III RPC4] |
0.464 |
-0.445 |
| 166 |
246733_at |
AT5G27660
|
DEG14, degradation of periplasmic proteins 14 |
0.462 |
0.048 |
| 167 |
264799_at |
AT1G08550
|
AVDE1, ARABIDOPSIS VIOLAXANTHIN DE-EPOXIDASE 1, NPQ1, non-photochemical quenching 1 |
0.462 |
-0.035 |
| 168 |
256797_at |
AT3G18600
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.462 |
0.335 |
| 169 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.460 |
0.011 |
| 170 |
245456_at |
AT4G16950
|
RPP5, RECOGNITION OF PERONOSPORA PARASITICA 5 |
0.460 |
0.082 |
| 171 |
263483_at |
AT2G04030
|
AtHsp90.5, HEAT SHOCK PROTEIN 90.5, AtHsp90C, CR88, EMB1956, EMBRYO DEFECTIVE 1956, Hsp88.1, HEAT SHOCK PROTEIN 88.1, HSP90.5, HEAT SHOCK PROTEIN 90.5 |
0.459 |
-0.121 |
| 172 |
252252_at |
AT3G49180
|
RID3, ROOT INITIATION DEFECTIVE 3 |
0.459 |
0.145 |
| 173 |
245743_at |
AT1G51080
|
unknown |
0.457 |
0.003 |
| 174 |
246262_at |
AT1G31790
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.457 |
0.230 |
| 175 |
251678_at |
AT3G56990
|
EDA7, embryo sac development arrest 7 |
0.455 |
-0.036 |
| 176 |
254303_at |
AT4G22830
|
unknown |
0.455 |
0.194 |
| 177 |
255465_at |
AT4G02990
|
BSM, BELAYA SMERT, RUG2, RUGOSA 2 |
0.453 |
-0.140 |
| 178 |
245712_at |
AT5G04360
|
ATLDA, limit dextrinase, ATPU1, PULLULANASE 1, LDA, limit dextrinase, PU1, PULLULANASE 1 |
0.451 |
0.083 |
| 179 |
260157_at |
AT1G52930
|
[Ribosomal RNA processing Brix domain protein] |
0.450 |
-0.009 |
| 180 |
259503_at |
AT1G15870
|
[Mitochondrial glycoprotein family protein] |
0.449 |
0.297 |
| 181 |
248890_at |
AT5G46270
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.449 |
0.087 |
| 182 |
263488_at |
AT2G31840
|
MRL7-L, Mesophyll-cell RNAi Library line 7-like |
0.447 |
0.007 |
| 183 |
259225_at |
AT3G07590
|
[Small nuclear ribonucleoprotein family protein] |
0.446 |
-0.107 |
| 184 |
256881_at |
AT3G26410
|
AtTRM11, TRM11, tRNA modification 11 |
0.445 |
-0.050 |
| 185 |
256837_at |
AT3G22900
|
NRPD7 |
0.444 |
-0.024 |
| 186 |
265596_at |
AT2G20020
|
ATCAF1, CAF1 |
0.444 |
-0.033 |
| 187 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.442 |
0.110 |
| 188 |
245188_at |
AT1G67660
|
[Restriction endonuclease, type II-like superfamily protein] |
0.442 |
0.018 |
| 189 |
266478_at |
AT2G31170
|
FIONA, FIONA, SYCO ARATH, cysteinyl t-RNA synthetase |
0.441 |
-0.132 |
| 190 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
0.439 |
0.146 |
| 191 |
245952_at |
AT5G28500
|
unknown |
0.438 |
0.042 |
| 192 |
260351_at |
AT1G69380
|
RRG, RETARDED ROOT GROWTH |
0.438 |
-0.322 |
| 193 |
245027_at |
AT2G26550
|
HO2, heme oxygenase 2 |
0.437 |
-0.050 |
| 194 |
266916_at |
AT2G45860
|
unknown |
0.436 |
-0.071 |
| 195 |
250198_at |
AT5G14100
|
ABCI11, ATP-binding cassette I11, ATNAP14, ARABIDOPSIS THALIANANON-INTRINSIC ABC PROTEIN 14, NAP14, non-intrinsic ABC protein 14 |
0.435 |
0.065 |
| 196 |
259422_at |
AT1G13810
|
[Restriction endonuclease, type II-like superfamily protein] |
0.435 |
-0.066 |
| 197 |
262168_at |
AT1G74730
|
unknown |
0.435 |
0.163 |
| 198 |
256796_at |
AT3G22210
|
unknown |
0.434 |
0.031 |
| 199 |
247415_at |
AT5G63060
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.434 |
0.102 |
| 200 |
245227_s_at |
AT1G08410
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.433 |
0.020 |
| 201 |
245670_at |
AT1G28210
|
ATJ1 |
0.433 |
0.008 |
| 202 |
260076_at |
AT1G73630
|
[EF hand calcium-binding protein family] |
0.432 |
0.136 |
| 203 |
254642_at |
AT4G18810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.432 |
0.152 |
| 204 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
0.432 |
0.052 |
| 205 |
263477_at |
AT2G31790
|
[UDP-Glycosyltransferase superfamily protein] |
0.431 |
0.153 |
| 206 |
256631_at |
AT3G28320
|
unknown |
0.431 |
0.068 |
| 207 |
260872_at |
AT1G21350
|
[Thioredoxin superfamily protein] |
0.429 |
-0.032 |
| 208 |
247420_at |
AT5G63100
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.428 |
0.155 |
| 209 |
263980_at |
AT2G42770
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.428 |
-0.186 |
| 210 |
253607_at |
AT4G30330
|
[Small nuclear ribonucleoprotein family protein] |
0.427 |
-0.059 |
| 211 |
246488_at |
AT5G16010
|
[3-oxo-5-alpha-steroid 4-dehydrogenase family protein] |
0.426 |
0.086 |
| 212 |
246057_at |
AT5G08400
|
unknown |
0.426 |
-0.035 |
| 213 |
263761_at |
AT2G21330
|
AtFBA1, FBA1, fructose-bisphosphate aldolase 1 |
0.426 |
0.071 |
| 214 |
246110_at |
AT5G20140
|
AtHBP5, HBP5, haem-binding protein 5 |
0.426 |
-0.051 |
| 215 |
247800_at |
AT5G58570
|
unknown |
0.426 |
-0.022 |
| 216 |
246814_at |
AT5G27200
|
ACP5, acyl carrier protein 5 |
0.425 |
0.108 |
| 217 |
262338_at |
AT1G64185
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
0.425 |
-0.028 |
| 218 |
258079_at |
AT3G25940
|
[TFIIB zinc-binding protein] |
0.423 |
-0.257 |
| 219 |
253252_at |
AT4G34740
|
ASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATPURF2, CIA1, CHLOROPLAST IMPORT APPARATUS 1, DOV1, DIFFERENTIAL DEVELOPMENT OF VASCULAR ASSOCIATED CELLS |
0.422 |
0.017 |
| 220 |
260125_at |
AT1G36390
|
[Co-chaperone GrpE family protein] |
0.422 |
-0.064 |
| 221 |
265959_at |
AT2G37240
|
[Thioredoxin superfamily protein] |
0.421 |
-0.017 |
| 222 |
260412_at |
AT1G69830
|
AMY3, alpha-amylase-like 3, ATAMY3, ALPHA-AMYLASE-LIKE 3 |
0.420 |
-0.133 |
| 223 |
247291_at |
AT5G64480
|
unknown |
0.420 |
-0.015 |
| 224 |
249966_at |
AT5G19030
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.419 |
0.087 |
| 225 |
248538_at |
AT5G50110
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.419 |
0.036 |
| 226 |
247247_at |
AT5G64650
|
[Ribosomal protein L17 family protein] |
0.418 |
-0.294 |
| 227 |
251814_at |
AT3G54890
|
LHCA1, photosystem I light harvesting complex gene 1 |
0.417 |
0.094 |
| 228 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
0.417 |
-0.189 |
| 229 |
263760_at |
AT2G21280
|
ATSULA, GC1, GIANT CHLOROPLAST 1, SULA |
0.417 |
-0.034 |
| 230 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.417 |
0.222 |
| 231 |
251371_at |
AT3G60360
|
EDA14, EMBRYO SAC DEVELOPMENT ARREST 14, UTP11, U3 SMALL NUCLEOLAR RNA-ASSOCIATED PROTEIN 11 |
0.417 |
-0.000 |
| 232 |
248173_at |
AT5G54580
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.417 |
-0.100 |
| 233 |
267174_at |
AT2G37600
|
[Ribosomal protein L36e family protein] |
0.416 |
-0.028 |
| 234 |
247649_at |
AT5G60030
|
unknown |
0.415 |
-0.071 |
| 235 |
247015_at |
AT5G66960
|
[Prolyl oligopeptidase family protein] |
0.415 |
0.085 |
| 236 |
246517_at |
AT5G15760
|
PSRP3/2, plastid-specific ribosomal protein 3/2 |
0.415 |
-0.041 |
| 237 |
250146_at |
AT5G14660
|
ATDEF2, DEF2, PDF1B, peptide deformylase 1B |
0.414 |
-0.154 |
| 238 |
258782_at |
AT3G11750
|
FOLB1 |
0.414 |
0.263 |
| 239 |
251611_at |
AT3G57940
|
unknown |
0.414 |
-0.034 |
| 240 |
252508_at |
AT3G46210
|
[Ribosomal protein S5 domain 2-like superfamily protein] |
0.413 |
-0.065 |
| 241 |
256349_at |
AT1G54890
|
[Late embryogenesis abundant (LEA) protein-related] |
0.413 |
0.060 |
| 242 |
255227_at |
AT4G05440
|
EDA35, embryo sac development arrest 35 |
0.413 |
-0.202 |
| 243 |
253689_at |
AT4G29770
|
unknown |
0.412 |
0.013 |
| 244 |
251355_at |
AT3G61100
|
[Putative endonuclease or glycosyl hydrolase] |
0.410 |
0.135 |
| 245 |
264738_at |
AT1G62250
|
unknown |
0.410 |
0.087 |
| 246 |
255440_at |
AT4G02530
|
[chloroplast thylakoid lumen protein] |
0.410 |
0.021 |
| 247 |
246746_at |
AT5G27820
|
[Ribosomal L18p/L5e family protein] |
0.410 |
0.194 |
| 248 |
258034_at |
AT3G21300
|
[RNA methyltransferase family protein] |
0.409 |
0.080 |
| 249 |
257909_at |
AT3G25480
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.408 |
0.014 |
| 250 |
249932_at |
AT5G22390
|
unknown |
0.408 |
0.038 |
| 251 |
265895_at |
AT2G15000
|
unknown |
0.407 |
-0.099 |
| 252 |
245765_at |
AT1G33600
|
[Leucine-rich repeat (LRR) family protein] |
0.406 |
-0.038 |
| 253 |
254645_at |
AT4G18520
|
SEL1, SEEDLING LETHAL 1 |
0.406 |
-0.245 |
| 254 |
264959_at |
AT1G77090
|
[Mog1/PsbP/DUF1795-like photosystem II reaction center PsbP family protein] |
0.405 |
-0.109 |
| 255 |
257903_at |
AT3G28460
|
[methyltransferases] |
0.404 |
0.036 |
| 256 |
249464_at |
AT5G39710
|
EMB2745, EMBRYO DEFECTIVE 2745 |
0.403 |
0.244 |
| 257 |
252001_at |
AT3G52750
|
FTSZ2-2 |
0.402 |
-0.270 |
| 258 |
255035_at |
AT4G09550
|
ATGIP1, ARABIDOPSIS ATGCP3 INTERACTING PROTEIN 1, GIP1, AtGCP3 interacting protein 1, GIP1a, GCP3-Interacting Protein 1a |
0.402 |
0.001 |
| 259 |
262609_at |
AT1G13930
|
unknown |
0.402 |
-0.185 |
| 260 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
0.400 |
0.088 |
| 261 |
250073_at |
AT5G17170
|
ENH1, enhancer of sos3-1 |
0.400 |
0.129 |
| 262 |
258161_at |
AT3G17930
|
DAC, Defective Accumulation of Cytochrome b6/f complex |
0.400 |
-0.028 |
| 263 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
0.400 |
0.052 |
| 264 |
251243_at |
AT3G61870
|
unknown |
0.399 |
0.092 |
| 265 |
267474_at |
AT2G02740
|
ATWHY3, A. THALIANA WHIRLY 3, PTAC11, PLASTID TRANSCRIPTIONALLY ACTIVE11, WHY3, WHIRLY 3 |
0.399 |
0.166 |
| 266 |
254504_at |
AT4G20030
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.399 |
0.153 |
| 267 |
255888_at |
AT1G20300
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.397 |
-0.140 |
| 268 |
247235_at |
AT5G64580
|
EMB3144, EMBRYO DEFECTIVE 3144 |
0.396 |
-0.172 |
| 269 |
258534_at |
AT3G06730
|
TRX P, thioredoxin putative plastidic, TRX z, Thioredoxin z |
0.396 |
-0.083 |
| 270 |
245901_at |
AT5G11060
|
KNAT4, KNOTTED1-like homeobox gene 4 |
0.396 |
0.653 |
| 271 |
245512_at |
AT4G15770
|
[RNA binding] |
0.394 |
0.015 |
| 272 |
247400_at |
AT5G62840
|
[Phosphoglycerate mutase family protein] |
0.393 |
0.071 |
| 273 |
252946_at |
AT4G39235
|
unknown |
0.393 |
-0.012 |
| 274 |
260601_at |
AT1G55910
|
ZIP11, zinc transporter 11 precursor |
0.393 |
-0.111 |
| 275 |
257188_at |
AT3G13150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.393 |
0.176 |
| 276 |
253740_at |
AT4G28706
|
[pfkB-like carbohydrate kinase family protein] |
0.393 |
0.107 |
| 277 |
265073_at |
AT1G55480
|
ZKT, protein containing PDZ domain, a K-box domain, and a TPR region |
0.391 |
-0.078 |
| 278 |
263676_at |
AT1G09340
|
CRB, chloroplast RNA binding, CSP41B, CHLOROPLAST STEM-LOOP BINDING PROTEIN OF 41 KDA, HIP1.3, heteroglycan-interacting protein 1.3 |
0.391 |
0.205 |
| 279 |
262738_at |
AT1G28530
|
unknown |
0.391 |
-0.145 |
| 280 |
260824_at |
AT1G06720
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.391 |
-0.214 |
| 281 |
261141_at |
AT1G19740
|
[ATP-dependent protease La (LON) domain protein] |
0.390 |
0.123 |
| 282 |
256786_at |
AT3G13740
|
[Ribonuclease III family protein] |
0.390 |
-0.030 |
| 283 |
248473_at |
AT5G50810
|
TIM8, translocase inner membrane subunit 8 |
0.389 |
-0.111 |
| 284 |
265139_at |
AT1G51310
|
[transferases] |
0.389 |
-0.042 |
| 285 |
262507_at |
AT1G11330
|
[S-locus lectin protein kinase family protein] |
0.388 |
0.239 |
| 286 |
251157_at |
AT3G63140
|
CSP41A, chloroplast stem-loop binding protein of 41 kDa |
0.388 |
0.073 |
| 287 |
266767_at |
AT2G46910
|
[Plastid-lipid associated protein PAP / fibrillin family protein] |
0.388 |
0.175 |
| 288 |
262669_at |
AT1G62850
|
[Class I peptide chain release factor] |
0.388 |
-0.034 |
| 289 |
253197_at |
AT4G35250
|
HCF244, high chlorophyll fluorescence phenotype 244 |
0.387 |
0.006 |
| 290 |
261048_at |
AT1G01420
|
UGT72B3, UDP-glucosyl transferase 72B3 |
0.387 |
0.125 |
| 291 |
256076_at |
AT1G18060
|
unknown |
0.387 |
0.049 |
| 292 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
0.387 |
0.065 |
| 293 |
265704_at |
AT2G03420
|
unknown |
0.386 |
-0.090 |
| 294 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.386 |
-0.043 |
| 295 |
262941_at |
AT1G79490
|
EMB2217, embryo defective 2217 |
0.385 |
0.273 |
| 296 |
248984_at |
AT5G45140
|
NRPC2, nuclear RNA polymerase C2 |
0.385 |
-0.349 |
| 297 |
259347_at |
AT3G03920
|
[H/ACA ribonucleoprotein complex, subunit Gar1/Naf1 protein] |
0.385 |
-0.219 |
| 298 |
247616_at |
AT5G60260
|
unknown |
0.385 |
0.282 |
| 299 |
248028_at |
AT5G55620
|
unknown |
0.385 |
0.170 |
| 300 |
267430_at |
AT2G34860
|
EDA3, embryo sac development arrest 3 |
0.385 |
-0.021 |
| 301 |
248944_at |
AT5G45500
|
[RNI-like superfamily protein] |
-1.392 |
0.014 |
| 302 |
256021_at |
AT1G58270
|
ZW9 |
-1.381 |
-0.246 |
| 303 |
255160_at |
AT4G07820
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-1.318 |
0.046 |
| 304 |
249053_at |
AT5G44440
|
[FAD-binding Berberine family protein] |
-1.313 |
0.039 |
| 305 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-1.290 |
0.140 |
| 306 |
245970_at |
AT5G20710
|
BGAL7, beta-galactosidase 7 |
-1.141 |
0.262 |
| 307 |
250165_at |
AT5G15290
|
CASP5, Casparian strip membrane domain protein 5 |
-1.139 |
0.235 |
| 308 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
-1.089 |
0.299 |
| 309 |
262206_at |
AT2G01090
|
[Ubiquinol-cytochrome C reductase hinge protein] |
-1.078 |
0.056 |
| 310 |
256712_at |
AT2G34020
|
[Calcium-binding EF-hand family protein] |
-1.061 |
0.192 |
| 311 |
255805_at |
AT4G10240
|
bbx23, B-box domain protein 23 |
-1.009 |
0.114 |
| 312 |
248435_at |
AT5G51210
|
OLEO3, oleosin3 |
-1.006 |
0.127 |
| 313 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
-1.004 |
-0.005 |
| 314 |
263276_at |
AT2G14100
|
CYP705A13, cytochrome P450, family 705, subfamily A, polypeptide 13 |
-0.995 |
0.051 |
| 315 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-0.967 |
0.212 |
| 316 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.953 |
0.073 |
| 317 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.925 |
0.289 |
| 318 |
248213_at |
AT5G53660
|
AtGRF7, growth-regulating factor 7, GRF7, growth-regulating factor 7 |
-0.887 |
0.140 |
| 319 |
255546_at |
AT4G01910
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.886 |
0.209 |
| 320 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
-0.881 |
-0.211 |
| 321 |
249205_at |
AT5G42600
|
MRN1, marneral synthase 1 |
-0.862 |
-0.077 |
| 322 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
-0.858 |
-0.102 |
| 323 |
249814_at |
AT5G23840
|
[MD-2-related lipid recognition domain-containing protein] |
-0.857 |
-0.061 |
| 324 |
255484_at |
AT4G02540
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.843 |
-0.046 |
| 325 |
263894_at |
AT2G21910
|
CYP96A5, cytochrome P450, family 96, subfamily A, polypeptide 5 |
-0.842 |
0.160 |
| 326 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-0.836 |
-0.243 |
| 327 |
257689_at |
AT3G12820
|
AtMYB10, myb domain protein 10, MYB10, myb domain protein 10 |
-0.834 |
0.171 |
| 328 |
248079_at |
AT5G55790
|
unknown |
-0.825 |
-0.031 |
| 329 |
267264_at |
AT2G22970
|
SCPL11, serine carboxypeptidase-like 11 |
-0.822 |
0.102 |
| 330 |
254876_at |
AT4G11610
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.813 |
0.360 |
| 331 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.808 |
0.123 |
| 332 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.804 |
0.189 |
| 333 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.800 |
-0.071 |
| 334 |
247159_at |
AT5G65800
|
ACS5, ACC synthase 5, ATACS5, CIN5, CYTOKININ-INSENSITIVE 5, ETO2, ETHYLENE OVERPRODUCER 2 |
-0.797 |
0.063 |
| 335 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.796 |
-0.037 |
| 336 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.790 |
0.277 |
| 337 |
254327_at |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.778 |
0.198 |
| 338 |
266688_at |
AT2G19660
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.771 |
0.082 |
| 339 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.769 |
0.203 |
| 340 |
248245_at |
AT5G53190
|
AtSWEET3, SWEET3 |
-0.768 |
-0.024 |
| 341 |
255851_at |
AT1G67040
|
TRM22, TON1 Recruiting Motif 22 |
-0.760 |
0.164 |
| 342 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.742 |
0.070 |
| 343 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
-0.737 |
-0.335 |
| 344 |
262121_at |
AT1G02800
|
ATCEL2, cellulase 2, CEL2, CEL2, cellulase 2 |
-0.735 |
0.240 |
| 345 |
246888_at |
AT5G26270
|
unknown |
-0.734 |
-0.053 |
| 346 |
246526_at |
AT5G15720
|
GLIP7, GDSL-motif lipase 7 |
-0.727 |
-0.159 |
| 347 |
254313_at |
AT4G22460
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.726 |
0.181 |
| 348 |
262458_at |
AT1G11280
|
[S-locus lectin protein kinase family protein] |
-0.724 |
-0.074 |
| 349 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.722 |
-0.607 |
| 350 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
-0.721 |
0.113 |
| 351 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.720 |
0.005 |
| 352 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
-0.718 |
0.227 |
| 353 |
255608_at |
AT4G01140
|
unknown |
-0.711 |
0.504 |
| 354 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-0.706 |
0.287 |
| 355 |
248978_at |
AT5G45070
|
AtPP2-A8, phloem protein 2-A8, PP2-A8, phloem protein 2-A8 |
-0.692 |
0.271 |
| 356 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
-0.681 |
0.263 |
| 357 |
250597_at |
AT5G07680
|
ANAC079, Arabidopsis NAC domain containing protein 79, ANAC080, NAC domain containing protein 80, ATNAC4, NAC080, NAC domain containing protein 80, NAC4 |
-0.679 |
0.116 |
| 358 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
-0.669 |
-0.094 |
| 359 |
262421_at |
AT1G50290
|
unknown |
-0.668 |
-0.116 |
| 360 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
-0.667 |
-0.146 |
| 361 |
265345_at |
AT2G22680
|
WAVH1, WAV3 homolog 1 |
-0.664 |
-0.138 |
| 362 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
-0.659 |
0.105 |
| 363 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
-0.652 |
-0.090 |
| 364 |
261224_at |
AT1G20160
|
ATSBT5.2 |
-0.648 |
-0.013 |
| 365 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.647 |
0.270 |
| 366 |
249614_at |
AT5G37300
|
WSD1 |
-0.647 |
0.139 |
| 367 |
246393_at |
AT1G58150
|
unknown |
-0.644 |
0.024 |
| 368 |
248416_at |
AT5G51630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.638 |
0.093 |
| 369 |
266222_at |
AT2G28780
|
unknown |
-0.637 |
-0.097 |
| 370 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.636 |
0.102 |
| 371 |
253182_at |
AT4G35190
|
LOG5, LONELY GUY 5 |
-0.633 |
0.190 |
| 372 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
-0.622 |
0.037 |
| 373 |
255463_at |
AT4G02960
|
ATRE2, retro element 2, RE2, retro element 2 |
-0.621 |
0.171 |
| 374 |
265296_at |
AT2G14060
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.620 |
0.081 |
| 375 |
248415_at |
AT5G51620
|
[Uncharacterised protein family (UPF0172)] |
-0.620 |
0.059 |
| 376 |
260004_at |
AT1G67860
|
unknown |
-0.613 |
0.211 |
| 377 |
266753_at |
AT2G46990
|
IAA20, indole-3-acetic acid inducible 20 |
-0.613 |
-0.037 |
| 378 |
266978_at |
AT2G39430
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.606 |
0.190 |
| 379 |
252458_at |
AT3G47210
|
unknown |
-0.604 |
-0.117 |
| 380 |
247868_at |
AT5G57620
|
AtMYB36, myb domain protein 36, MYB36, myb domain protein 36 |
-0.600 |
0.168 |
| 381 |
259799_at |
AT1G72290
|
[Kunitz family trypsin and protease inhibitor protein] |
-0.595 |
0.021 |
| 382 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
-0.592 |
0.058 |
| 383 |
265645_at |
AT2G27370
|
CASP3, Casparian strip membrane domain protein 3 |
-0.587 |
0.153 |
| 384 |
267414_at |
AT2G34790
|
EDA28, EMBRYO SAC DEVELOPMENT ARREST 28, MEE23, MATERNAL EFFECT EMBRYO ARREST 23 |
-0.587 |
0.002 |
| 385 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.587 |
0.098 |
| 386 |
257113_at |
AT3G20130
|
CYP705A22, cytochrome P450, family 705, subfamily A, polypeptide 22, GPS1, gravity persistence signal 1 |
-0.586 |
-0.264 |
| 387 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
-0.582 |
0.587 |
| 388 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.579 |
-0.009 |
| 389 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.577 |
-0.109 |
| 390 |
245410_at |
AT4G17220
|
ATMAP70-5, microtubule-associated proteins 70-5, MAP70-5, microtubule-associated proteins 70-5 |
-0.576 |
0.130 |
| 391 |
267160_at |
AT2G37670
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.576 |
0.112 |
| 392 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.575 |
0.065 |
| 393 |
251116_at |
AT3G63470
|
scpl40, serine carboxypeptidase-like 40 |
-0.575 |
0.015 |
| 394 |
247360_at |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
-0.574 |
-0.183 |
| 395 |
260181_at |
AT1G70710
|
ATGH9B1, glycosyl hydrolase 9B1, CEL1, CELLULASE 1, GH9B1, glycosyl hydrolase 9B1 |
-0.573 |
0.154 |
| 396 |
249439_at |
AT5G40020
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.571 |
0.105 |
| 397 |
253054_at |
AT4G37580
|
COP3, CONSTITUTIVE PHOTOMORPHOGENIC 3, HLS1, HOOKLESS 1, UNS2, UNUSUAL SUGAR RESPONSE 2 |
-0.571 |
-0.007 |
| 398 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
-0.570 |
-0.198 |
| 399 |
266356_at |
AT2G32300
|
UCC1, uclacyanin 1 |
-0.569 |
0.272 |
| 400 |
265246_at |
AT2G43050
|
ATPMEPCRD |
-0.569 |
0.154 |
| 401 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
-0.567 |
0.436 |
| 402 |
260664_at |
AT1G19510
|
ATRL5, RAD-like 5, RL5, RAD-like 5, RSM4, RADIALIS-LIKE SANT/MYB 4 |
-0.560 |
0.146 |
| 403 |
247582_at |
AT5G60760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.558 |
0.211 |
| 404 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
-0.557 |
0.098 |
| 405 |
245439_at |
AT4G16670
|
unknown |
-0.556 |
0.209 |
| 406 |
260074_at |
AT1G73640
|
AtRABA6a, RAB GTPase homolog A6A, RABA6a, RAB GTPase homolog A6A |
-0.554 |
-0.012 |
| 407 |
259291_at |
AT3G11550
|
CASP2, Casparian strip membrane domain protein 2 |
-0.550 |
-0.012 |
| 408 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.550 |
0.090 |
| 409 |
250717_at |
AT5G06200
|
CASP4, Casparian strip membrane domain protein 4 |
-0.546 |
0.038 |
| 410 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-0.544 |
-0.052 |
| 411 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.544 |
-0.241 |
| 412 |
249033_at |
AT5G44920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.543 |
0.061 |
| 413 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
-0.543 |
-0.238 |
| 414 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.542 |
-0.045 |
| 415 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.541 |
0.618 |
| 416 |
259429_at |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
-0.541 |
0.141 |
| 417 |
250214_at |
AT5G13870
|
EXGT-A4, endoxyloglucan transferase A4, XTH5, xyloglucan endotransglucosylase/hydrolase 5 |
-0.538 |
0.038 |
| 418 |
247648_at |
AT5G60020
|
ATLAC17, LAC17, laccase 17 |
-0.537 |
0.100 |
| 419 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
-0.536 |
0.190 |
| 420 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.535 |
0.099 |
| 421 |
246138_at |
AT5G19870
|
unknown |
-0.535 |
0.212 |
| 422 |
248936_at |
AT5G45710
|
AT-HSFA4C, HEAT SHOCK TRANSCRIPTION FACTOR A4C, HSFA4C, HEAT SHOCK TRANSCRIPTION FACTOR A4C, RHA1, ROOT HANDEDNESS 1 |
-0.534 |
0.054 |
| 423 |
262756_at |
AT1G16370
|
ATOCT6, ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER 6, OCT6, organic cation/carnitine transporter 6 |
-0.534 |
0.084 |
| 424 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.533 |
0.127 |
| 425 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
-0.532 |
0.080 |
| 426 |
264395_at |
AT1G12070
|
[Immunoglobulin E-set superfamily protein] |
-0.531 |
0.029 |
| 427 |
257112_at |
AT3G20120
|
CYP705A21, cytochrome P450, family 705, subfamily A, polypeptide 21 |
-0.527 |
-0.163 |
| 428 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
-0.527 |
0.216 |
| 429 |
257022_at |
AT3G19580
|
AZF2, zinc-finger protein 2, ZF2, zinc-finger protein 2 |
-0.526 |
0.135 |
| 430 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
-0.526 |
0.198 |
| 431 |
267307_at |
AT2G30210
|
LAC3, laccase 3 |
-0.523 |
0.140 |
| 432 |
262198_at |
AT1G53830
|
ATPME2, pectin methylesterase 2, PME2, pectin methylesterase 2 |
-0.522 |
-0.053 |
| 433 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.518 |
0.135 |
| 434 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
-0.517 |
0.128 |
| 435 |
257899_at |
AT3G28345
|
ABCB15, ATP-binding cassette B15, MDR13, multi-drug resistance 13 |
-0.517 |
-0.199 |
| 436 |
247586_at |
AT5G60660
|
PIP2;4, plasma membrane intrinsic protein 2;4, PIP2F |
-0.517 |
0.266 |
| 437 |
262918_at |
AT1G65000
|
unknown |
-0.516 |
0.010 |
| 438 |
262485_at |
AT1G21730
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.514 |
-0.057 |
| 439 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-0.511 |
0.010 |
| 440 |
245550_at |
AT4G15330
|
CYP705A1, cytochrome P450, family 705, subfamily A, polypeptide 1 |
-0.511 |
0.178 |
| 441 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
-0.509 |
0.078 |
| 442 |
254506_at |
AT4G20140
|
GSO1, GASSHO1 |
-0.508 |
0.307 |
| 443 |
247951_at |
AT5G57240
|
ORP4C, OSBP(oxysterol binding protein)-related protein 4C |
-0.507 |
-0.072 |
| 444 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
-0.507 |
-0.117 |
| 445 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.505 |
-0.026 |
| 446 |
245903_at |
AT5G11100
|
ATSYTD, NTMC2T2.2, NTMC2TYPE2.2, SYT4, synaptotagmin 4, SYTD |
-0.504 |
0.075 |
| 447 |
247020_at |
AT5G67020
|
unknown |
-0.503 |
-0.025 |
| 448 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.503 |
0.028 |
| 449 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
-0.501 |
0.068 |
| 450 |
256114_at |
AT1G16850
|
unknown |
-0.500 |
-0.137 |
| 451 |
266783_at |
AT2G29130
|
ATLAC2, LAC2, laccase 2 |
-0.498 |
0.090 |
| 452 |
259274_at |
AT3G01220
|
ATHB20, homeobox protein 20, HB20, homeobox protein 20 |
-0.498 |
0.114 |
| 453 |
251517_at |
AT3G59370
|
[Vacuolar calcium-binding protein-related] |
-0.497 |
0.080 |
| 454 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.496 |
0.068 |
| 455 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.496 |
0.009 |
| 456 |
264835_at |
AT1G03550
|
AtSCAMP4, SCAMP4, Secretory carrier membrane protein 4 |
-0.496 |
0.004 |
| 457 |
260841_at |
AT1G29195
|
unknown |
-0.495 |
0.036 |
| 458 |
255348_at |
AT4G03820
|
unknown |
-0.494 |
-0.069 |
| 459 |
253096_at |
AT4G37330
|
CYP81D4, cytochrome P450, family 81, subfamily D, polypeptide 4 |
-0.494 |
0.203 |
| 460 |
250771_at |
AT5G05400
|
[LRR and NB-ARC domains-containing disease resistance protein] |
-0.492 |
0.049 |
| 461 |
250697_at |
AT5G06800
|
[myb-like HTH transcriptional regulator family protein] |
-0.492 |
0.171 |
| 462 |
251768_at |
AT3G55940
|
[Phosphoinositide-specific phospholipase C family protein] |
-0.490 |
-0.152 |
| 463 |
247345_at |
AT5G63760
|
ARI15, ARIADNE 15, ATARI15, ARABIDOPSIS ARIADNE 15 |
-0.489 |
0.124 |
| 464 |
262362_at |
AT1G72840
|
[Disease resistance protein (TIR-NBS-LRR class)] |
-0.484 |
0.092 |
| 465 |
261247_at |
AT1G20070
|
unknown |
-0.482 |
0.162 |
| 466 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
-0.482 |
0.057 |
| 467 |
258935_at |
AT3G10120
|
unknown |
-0.480 |
-0.091 |
| 468 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
-0.477 |
-0.082 |
| 469 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.476 |
0.053 |
| 470 |
255858_at |
AT1G67030
|
ZFP6, zinc finger protein 6 |
-0.474 |
0.011 |
| 471 |
249227_at |
AT5G42180
|
PER64, peroxidase 64 |
-0.473 |
0.103 |
| 472 |
265688_at |
AT2G24300
|
[Calmodulin-binding protein] |
-0.471 |
0.028 |
| 473 |
250213_at |
AT5G13820
|
ATBP-1, ATBP1, ATTBP1, HPPBF-1, H-PROTEIN PROMOTE, TBP1, telomeric DNA binding protein 1 |
-0.470 |
0.180 |
| 474 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
-0.469 |
-0.037 |
| 475 |
246877_at |
AT5G26150
|
[protein kinase family protein] |
-0.469 |
0.120 |
| 476 |
262575_at |
AT1G15210
|
ABCG35, ATP-binding cassette G35, ATPDR7, PLEIOTROPIC DRUG RESISTANCE 7, PDR7, pleiotropic drug resistance 7 |
-0.469 |
0.206 |
| 477 |
249849_at |
AT5G23230
|
NIC2, nicotinamidase 2 |
-0.468 |
0.071 |
| 478 |
246197_at |
AT4G37010
|
CEN2, centrin 2 |
-0.466 |
0.029 |
| 479 |
259734_at |
AT1G77500
|
unknown |
-0.463 |
0.095 |
| 480 |
260220_at |
AT1G74650
|
ATMYB31, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 31, ATY13, MYB31, myb domain protein 31 |
-0.462 |
0.063 |
| 481 |
265261_at |
AT2G42990
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.462 |
0.311 |
| 482 |
247312_at |
AT5G63970
|
RGLG3, RING DOMAIN LIGASE 3 |
-0.461 |
0.204 |
| 483 |
266362_at |
AT2G32430
|
[Galactosyltransferase family protein] |
-0.461 |
0.138 |
| 484 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.459 |
0.231 |
| 485 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
-0.459 |
0.161 |
| 486 |
252400_at |
AT3G48020
|
unknown |
-0.458 |
0.059 |
| 487 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
-0.457 |
0.183 |
| 488 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
-0.456 |
-0.167 |
| 489 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
-0.456 |
-0.091 |
| 490 |
248579_at |
AT5G49880
|
AtMAD1, MAD1, mitotic arrest deficient 1, NES1, NITRIC OXIDE-INDUCED EARLY COTYLEDON SENESCENCE 1 |
-0.456 |
0.164 |
| 491 |
255702_at |
AT4G00230
|
XSP1, xylem serine peptidase 1 |
-0.455 |
0.090 |
| 492 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.455 |
-0.159 |
| 493 |
251870_at |
AT3G54510
|
[Early-responsive to dehydration stress protein (ERD4)] |
-0.454 |
0.447 |
| 494 |
249055_at |
AT5G44460
|
CML43, calmodulin like 43 |
-0.452 |
0.087 |
| 495 |
256721_at |
AT2G34150
|
ATRANGAP2, ATSCAR1, SCAR1, WAVE1, WISKOTT-ALDRICH SYNDROME PROTEIN FAMILY VERPROLIN HOMOLOGOUS PROTEIN 1 |
-0.452 |
0.110 |
| 496 |
254404_at |
AT4G21340
|
B70 |
-0.452 |
0.057 |
| 497 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
-0.451 |
0.214 |
| 498 |
266975_at |
AT2G39380
|
ATEXO70H2, exocyst subunit exo70 family protein H2, EXO70H2, exocyst subunit exo70 family protein H2 |
-0.451 |
0.379 |
| 499 |
251050_at |
AT5G02440
|
unknown |
-0.451 |
0.105 |
| 500 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
-0.450 |
0.268 |
| 501 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
-0.449 |
-0.100 |
| 502 |
254718_at |
AT4G13580
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.449 |
-0.006 |
| 503 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.446 |
0.139 |
| 504 |
262831_at |
AT1G14730
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
-0.445 |
0.072 |
| 505 |
253556_at |
AT4G31100
|
[wall-associated kinase, putative] |
-0.444 |
-0.190 |
| 506 |
254292_at |
AT4G23030
|
[MATE efflux family protein] |
-0.442 |
0.031 |
| 507 |
254836_at |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
-0.441 |
0.409 |
| 508 |
251023_at |
AT5G02170
|
[Transmembrane amino acid transporter family protein] |
-0.441 |
0.193 |
| 509 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
-0.439 |
0.020 |
| 510 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
-0.439 |
0.191 |
| 511 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.438 |
-0.235 |
| 512 |
265188_at |
AT1G23800
|
ALDH2B, aldehyde dehydrogenase 2B, ALDH2B7, aldehyde dehydrogenase 2B7 |
-0.438 |
-0.082 |
| 513 |
248424_at |
AT5G51680
|
[hydroxyproline-rich glycoprotein family protein] |
-0.438 |
-0.039 |
| 514 |
264184_at |
AT1G54790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.436 |
-0.009 |
| 515 |
255811_at |
AT4G10250
|
ATHSP22.0 |
-0.436 |
0.137 |
| 516 |
256425_at |
AT1G33560
|
ADR1, ACTIVATED DISEASE RESISTANCE 1 |
-0.434 |
0.046 |
| 517 |
251191_at |
AT3G62590
|
[alpha/beta-Hydrolases superfamily protein] |
-0.431 |
0.165 |
| 518 |
248697_at |
AT5G48370
|
[Thioesterase/thiol ester dehydrase-isomerase superfamily protein] |
-0.431 |
0.000 |
| 519 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
-0.429 |
0.837 |
| 520 |
247933_at |
AT5G56980
|
unknown |
-0.428 |
0.118 |
| 521 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.428 |
0.130 |
| 522 |
252447_at |
AT3G47040
|
[Glycosyl hydrolase family protein] |
-0.428 |
0.266 |
| 523 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
-0.428 |
0.340 |
| 524 |
250467_at |
AT5G10100
|
TPPI, trehalose-6-phosphate phosphatase I |
-0.425 |
0.184 |
| 525 |
247170_at |
AT5G65530
|
[Protein kinase superfamily protein] |
-0.425 |
0.266 |
| 526 |
251478_at |
AT3G59690
|
IQD13, IQ-domain 13 |
-0.424 |
0.180 |
| 527 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
-0.421 |
0.015 |
| 528 |
251289_at |
AT3G61830
|
ARF18, auxin response factor 18 |
-0.420 |
0.090 |
| 529 |
250103_at |
AT5G16600
|
AtMYB43, myb domain protein 43, MYB43, myb domain protein 43 |
-0.419 |
-0.005 |
| 530 |
265985_at |
AT2G24220
|
ATPUP5, purine permease 5, PUP5, purine permease 5 |
-0.419 |
0.076 |
| 531 |
255443_at |
AT4G02700
|
SULTR3;2, sulfate transporter 3;2 |
-0.418 |
0.180 |
| 532 |
249245_at |
AT5G42280
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.418 |
0.068 |
| 533 |
246374_at |
AT1G51840
|
[protein kinase-related] |
-0.417 |
0.348 |
| 534 |
265863_at |
AT2G01770
|
ATVIT1, VIT1, vacuolar iron transporter 1 |
-0.417 |
0.101 |
| 535 |
256621_at |
AT3G24450
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.417 |
-0.185 |
| 536 |
249185_at |
AT5G43030
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.417 |
0.038 |
| 537 |
263284_at |
AT2G36100
|
CASP1, Casparian strip membrane domain protein 1 |
-0.417 |
0.156 |
| 538 |
265131_at |
AT1G23760
|
JP630, PG3, POLYGALACTURONASE 3 |
-0.415 |
-0.045 |
| 539 |
265144_at |
AT1G51170
|
AGC2-3, AGC2 kinase 3, UCN, UNICORN |
-0.415 |
-0.008 |
| 540 |
255528_at |
AT4G02090
|
unknown |
-0.414 |
0.076 |
| 541 |
263470_at |
AT2G31900
|
ATMYO5, MYOSIN 5, ATXIF, MYOSIN XI F, XIF, myosin-like protein XIF |
-0.414 |
0.028 |
| 542 |
248977_at |
AT5G45020
|
[Glutathione S-transferase family protein] |
-0.413 |
0.032 |
| 543 |
260144_at |
AT1G71960
|
ABCG25, ATP-binding casette G25, ATABCG25, Arabidopsis thaliana ATP-binding cassette G25 |
-0.413 |
0.134 |
| 544 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
-0.412 |
0.045 |
| 545 |
247354_at |
AT5G63590
|
ATFLS3, FLS3, flavonol synthase 3 |
-0.411 |
0.145 |
| 546 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
-0.411 |
0.062 |
| 547 |
249870_at |
AT5G23080
|
TGH, TOUGH |
-0.411 |
-0.016 |
| 548 |
249136_at |
AT5G43180
|
unknown |
-0.410 |
-0.036 |
| 549 |
255547_at |
AT4G01920
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.410 |
0.322 |
| 550 |
253145_at |
AT4G35560
|
DAW1, DUO1-activated WD40 1 |
-0.409 |
-0.344 |
| 551 |
248824_at |
AT5G46940
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.409 |
0.158 |
| 552 |
260024_at |
AT1G30080
|
[Glycosyl hydrolase superfamily protein] |
-0.408 |
-0.019 |
| 553 |
267588_at |
AT2G42060
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.407 |
0.068 |
| 554 |
257618_at |
AT3G24715
|
[Protein kinase superfamily protein with octicosapeptide/Phox/Bem1p domain] |
-0.406 |
0.181 |
| 555 |
253011_at |
AT4G37890
|
EDA40, embryo sac development arrest 40 |
-0.405 |
0.061 |
| 556 |
254015_at |
AT4G26140
|
BGAL12, beta-galactosidase 12 |
-0.403 |
-0.079 |
| 557 |
248778_at |
AT5G47940
|
unknown |
-0.403 |
0.143 |
| 558 |
246425_at |
AT5G17420
|
ATCESA7, CESA7, CELLULOSE SYNTHASE CATALYTIC SUBUNIT 7, IRX3, IRREGULAR XYLEM 3, MUR10, MURUS 10 |
-0.402 |
0.106 |
| 559 |
262787_at |
AT1G10730
|
AP1M1, adaptor protein-1 mu-adaptin 1 |
-0.401 |
-0.091 |
| 560 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.401 |
0.065 |
| 561 |
259380_at |
AT3G16340
|
ABCG29, ATP-binding cassette G29, AtABCG29, ATPDR1, PLEIOTROPIC DRUG RESISTANCE 1, PDR1, pleiotropic drug resistance 1 |
-0.401 |
0.310 |
| 562 |
249946_at |
AT5G19170
|
unknown |
-0.400 |
-0.044 |
| 563 |
262444_at |
AT1G47480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.399 |
0.199 |
| 564 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
-0.399 |
0.277 |
| 565 |
257757_at |
AT3G18660
|
GUX1, glucuronic acid substitution of xylan 1, PGSIP1, plant glycogenin-like starch initiation protein 1 |
-0.398 |
0.083 |
| 566 |
247091_at |
AT5G66390
|
PRX72, PEROXIDASE 72 |
-0.398 |
0.329 |
| 567 |
245838_at |
AT1G58410
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.395 |
0.110 |
| 568 |
265245_at |
AT2G43060
|
AtIBH1, IBH1, ILI1 binding bHLH 1 |
-0.394 |
-0.101 |
| 569 |
262096_at |
AT1G56010
|
anac021, Arabidopsis NAC domain containing protein 21, ANAC022, Arabidopsis NAC domain containing protein 22, NAC1, NAC domain containing protein 1 |
-0.394 |
0.136 |
| 570 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.392 |
0.285 |
| 571 |
247126_at |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
-0.391 |
-0.177 |
| 572 |
257476_at |
AT1G80960
|
[F-box and Leucine Rich Repeat domains containing protein] |
-0.391 |
0.012 |
| 573 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
-0.391 |
0.105 |
| 574 |
256803_at |
AT3G20960
|
CYP705A33, cytochrome P450, family 705, subfamily A, polypeptide 33 |
-0.390 |
-0.059 |
| 575 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
-0.390 |
-0.105 |
| 576 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
-0.389 |
-0.022 |
| 577 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
-0.389 |
0.147 |
| 578 |
265302_at |
AT2G13970
|
[Mutator-like transposase family, has a 4.1e-39 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.389 |
0.088 |
| 579 |
259472_at |
AT1G18910
|
[zinc ion binding] |
-0.387 |
-0.046 |
| 580 |
263437_at |
AT2G28670
|
ESB1, ENHANCED SUBERIN 1 |
-0.387 |
-0.151 |
| 581 |
253842_at |
AT4G27860
|
MEB1, MEMBRANE OF ER BODY 1 |
-0.387 |
-0.153 |
| 582 |
247522_at |
AT5G61340
|
unknown |
-0.386 |
-0.017 |
| 583 |
264933_at |
AT1G61160
|
unknown |
-0.386 |
0.055 |
| 584 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.386 |
-0.033 |
| 585 |
247087_at |
AT5G66330
|
[Leucine-rich repeat (LRR) family protein] |
-0.386 |
0.194 |
| 586 |
252646_at |
AT3G44610
|
[Protein kinase superfamily protein] |
-0.385 |
-0.064 |
| 587 |
251769_at |
AT3G55950
|
ATCRR3, CCR3, CRINKLY4 related 3 |
-0.383 |
0.124 |
| 588 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.383 |
-0.065 |
| 589 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
-0.383 |
0.195 |
| 590 |
247590_at |
AT5G60720
|
unknown |
-0.382 |
0.041 |
| 591 |
266456_at |
AT2G22770
|
NAI1 |
-0.382 |
-0.101 |
| 592 |
251103_at |
AT5G01700
|
[Protein phosphatase 2C family protein] |
-0.382 |
0.009 |
| 593 |
254226_at |
AT4G23690
|
AtDIR6, Arabidopsis thaliana dirigent protein 6, DIR6, dirigent protein 6 |
-0.382 |
0.088 |
| 594 |
262244_at |
AT1G48260
|
CIPK17, CBL-interacting protein kinase 17, SnRK3.21, SNF1-RELATED PROTEIN KINASE 3.21 |
-0.381 |
0.359 |
| 595 |
253434_at |
AT4G32500
|
AKT5, K+ transporter 5, KT5, K+ transporter 5 |
-0.380 |
-0.009 |
| 596 |
266170_at |
AT2G39050
|
ArathEULS3, EULS3, Euonymus lectin S3 |
-0.380 |
-0.053 |
| 597 |
260966_at |
AT1G12220
|
RPS5, RESISTANT TO P. SYRINGAE 5 |
-0.379 |
-0.319 |
| 598 |
265031_at |
AT1G61590
|
[Protein kinase superfamily protein] |
-0.379 |
0.417 |
| 599 |
248423_at |
AT5G51670
|
unknown |
-0.378 |
-0.102 |
| 600 |
253580_at |
AT4G30400
|
[RING/U-box superfamily protein] |
-0.376 |
-0.022 |