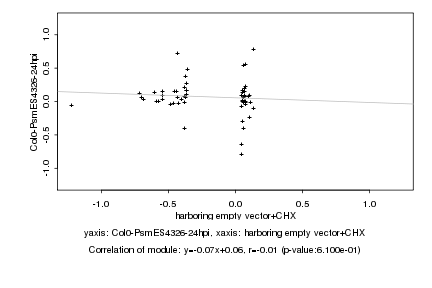
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
harboring empty vector+CHX |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
261429_at |
AT1G18860
|
ATWRKY61, WRKY DNA-BINDING PROTEIN 61, WRKY61, WRKY DNA-binding protein 61 |
0.133 |
0.785 |
| 2 |
265279_at |
AT2G28460
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.128 |
-0.102 |
| 3 |
264091_at |
AT1G79110
|
BRG2, BOI-related gene 2 |
0.108 |
-0.006 |
| 4 |
267128_at |
AT2G23620
|
ATMES1, ARABIDOPSIS THALIANA METHYL ESTERASE 1, MES1, methyl esterase 1 |
0.103 |
0.104 |
| 5 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
0.101 |
-0.228 |
| 6 |
260344_at |
AT1G69240
|
ATMES15, ARABIDOPSIS THALIANA METHYL ESTERASE 15, MES15, methyl esterase 15, RHS9, ROOT HAIR SPECIFIC 9 |
0.092 |
0.080 |
| 7 |
261780_at |
AT1G76310
|
CYCB2;4, CYCLIN B2;4 |
0.074 |
0.004 |
| 8 |
266776_at |
AT2G29070
|
[Ubiquitin fusion degradation UFD1 family protein] |
0.073 |
0.569 |
| 9 |
252751_at |
AT3G43430
|
[RING/U-box superfamily protein] |
0.068 |
-0.006 |
| 10 |
261280_at |
AT1G05860
|
unknown |
0.068 |
-0.041 |
| 11 |
255823_at |
AT2G40470
|
ASL11, ASYMMETRIC LEAVES2-LIKE 11, LBD15, LOB domain-containing protein 15 |
0.068 |
0.225 |
| 12 |
252758_at |
AT3G42710
|
[pseudogene] |
0.066 |
0.089 |
| 13 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
0.065 |
0.196 |
| 14 |
249882_at |
AT5G22890
|
STOP2, sensitive to proton rhizotoxicity 2 |
0.062 |
0.082 |
| 15 |
253499_at |
AT4G31900
|
CHR7, chromatin remodeling 7, PKR2, PICKLE RELATED 2 |
0.061 |
0.004 |
| 16 |
265871_at |
AT2G01680
|
[Ankyrin repeat family protein] |
0.061 |
0.162 |
| 17 |
251699_at |
AT3G56630
|
CYP94D2, cytochrome P450, family 94, subfamily D, polypeptide 2 |
0.061 |
0.096 |
| 18 |
251427_at |
AT3G60130
|
BGLU16, beta glucosidase 16 |
0.058 |
0.540 |
| 19 |
249986_at |
AT5G18460
|
unknown |
0.057 |
-0.403 |
| 20 |
263544_at |
AT2G21590
|
APL4 |
0.056 |
-0.002 |
| 21 |
246014_at |
AT5G10680
|
[calmodulin-binding protein-related] |
0.055 |
0.073 |
| 22 |
263814_at |
AT2G10010
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 0. P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
0.052 |
0.002 |
| 23 |
248237_at |
AT5G53890
|
AtPSKR2, PSKR2, phytosylfokine-alpha receptor 2 |
0.050 |
0.029 |
| 24 |
253712_at |
AT4G29330
|
DER1, DERLIN-1 |
0.049 |
0.179 |
| 25 |
259031_at |
AT3G09360
|
[Cyclin/Brf1-like TBP-binding protein] |
0.046 |
0.137 |
| 26 |
252829_at |
AT4G40060
|
ATHB-16, ATHB16, homeobox protein 16, HB16, homeobox protein 16 |
0.046 |
-0.299 |
| 27 |
255938_at |
AT1G12620
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.045 |
-0.060 |
| 28 |
255326_at |
AT4G04300
|
[pseudogene] |
0.044 |
0.103 |
| 29 |
266575_at |
AT2G24060
|
[Translation initiation factor 3 protein] |
0.043 |
-0.782 |
| 30 |
247191_at |
AT5G65310
|
ATHB-5, ATHB5, homeobox protein 5, HB5, homeobox protein 5 |
0.042 |
-0.640 |
| 31 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
-1.228 |
-0.048 |
| 32 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-0.718 |
0.121 |
| 33 |
259590_at |
AT1G28160
|
[Integrase-type DNA-binding superfamily protein] |
-0.705 |
0.075 |
| 34 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.688 |
0.034 |
| 35 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.611 |
0.141 |
| 36 |
245234_at |
AT4G25560
|
AtMYB18, myb domain protein 18, LAF1, LONG AFTER FAR-RED LIGHT 1, MYB18, myb domain protein 18 |
-0.594 |
0.012 |
| 37 |
264598_at |
AT1G04610
|
YUC3, YUCCA 3 |
-0.575 |
0.013 |
| 38 |
262981_at |
AT1G75590
|
SAUR52, SMALL AUXIN UPREGULATED RNA 52 |
-0.550 |
0.163 |
| 39 |
245113_at |
AT2G41660
|
MIZ1, mizu-kussei 1 |
-0.547 |
0.031 |
| 40 |
260607_at |
AT2G43700
|
LecRK-V.4, L-type lectin receptor kinase V.4 |
-0.545 |
0.103 |
| 41 |
256154_at |
AT3G08490
|
unknown |
-0.486 |
-0.044 |
| 42 |
245737_at |
AT1G44160
|
[HSP40/DnaJ peptide-binding protein] |
-0.467 |
-0.024 |
| 43 |
259576_at |
AT1G35330
|
[RING/U-box superfamily protein] |
-0.458 |
0.159 |
| 44 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
-0.444 |
0.159 |
| 45 |
258874_at |
AT3G03230
|
[alpha/beta-Hydrolases superfamily protein] |
-0.439 |
0.072 |
| 46 |
246844_at |
AT5G26660
|
ATMYB86, myb domain protein 86, MYB86, myb domain protein 86 |
-0.433 |
0.729 |
| 47 |
254692_at |
AT4G17860
|
unknown |
-0.425 |
-0.018 |
| 48 |
264425_at |
AT1G61750
|
[Receptor-like protein kinase-related family protein] |
-0.403 |
0.035 |
| 49 |
250231_at |
AT5G13910
|
LEP, LEAFY PETIOLE |
-0.386 |
-0.009 |
| 50 |
260039_at |
AT1G68795
|
CLE12, CLAVATA3/ESR-RELATED 12 |
-0.385 |
0.215 |
| 51 |
258196_at |
AT3G13980
|
unknown |
-0.384 |
-0.391 |
| 52 |
256847_at |
AT3G27950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.383 |
0.080 |
| 53 |
256788_at |
AT3G13730
|
CYP90D1, cytochrome P450, family 90, subfamily D, polypeptide 1 |
-0.375 |
0.376 |
| 54 |
250173_at |
AT5G14340
|
AtMYB40, myb domain protein 40, MYB40, myb domain protein 40 |
-0.374 |
0.072 |
| 55 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
-0.371 |
0.171 |
| 56 |
253527_at |
AT4G31470
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.368 |
0.171 |
| 57 |
265856_at |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
-0.368 |
0.113 |
| 58 |
266824_at |
AT2G22800
|
HAT9 |
-0.368 |
0.277 |
| 59 |
259596_at |
AT1G28130
|
GH3.17 |
-0.363 |
0.488 |