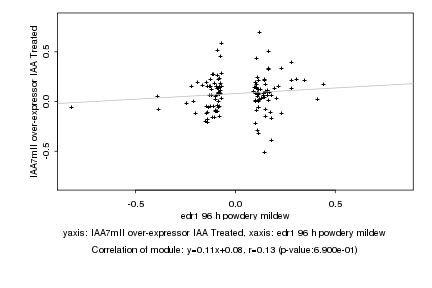
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
edr1 96 h powdery mildew |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.438 |
0.173 |
| 2 |
254574_at |
AT4G19430
|
unknown |
0.407 |
0.023 |
| 3 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.342 |
0.218 |
| 4 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.303 |
0.224 |
| 5 |
256382_at |
AT1G66860
|
[Class I glutamine amidotransferase-like superfamily protein] |
0.280 |
0.138 |
| 6 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.279 |
0.396 |
| 7 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.278 |
0.216 |
| 8 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
0.229 |
-0.119 |
| 9 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
0.228 |
0.340 |
| 10 |
256874_at |
AT3G26320
|
CYP71B36, cytochrome P450, family 71, subfamily B, polypeptide 36 |
0.211 |
0.155 |
| 11 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.202 |
0.035 |
| 12 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
0.191 |
0.141 |
| 13 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.179 |
-0.392 |
| 14 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.178 |
-0.168 |
| 15 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
0.178 |
0.068 |
| 16 |
266456_at |
AT2G22770
|
NAI1 |
0.172 |
-0.101 |
| 17 |
258225_at |
AT3G15630
|
unknown |
0.168 |
0.097 |
| 18 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.164 |
0.328 |
| 19 |
263182_at |
AT1G05575
|
unknown |
0.164 |
0.333 |
| 20 |
260656_at |
AT1G19380
|
unknown |
0.162 |
0.016 |
| 21 |
247324_at |
AT5G64190
|
unknown |
0.161 |
0.508 |
| 22 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.159 |
0.068 |
| 23 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.156 |
0.117 |
| 24 |
261924_at |
AT1G22550
|
[Major facilitator superfamily protein] |
0.156 |
0.092 |
| 25 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
0.150 |
0.102 |
| 26 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
0.149 |
0.091 |
| 27 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.148 |
-0.148 |
| 28 |
247212_at |
AT5G65040
|
unknown |
0.148 |
-0.075 |
| 29 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.146 |
0.173 |
| 30 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.143 |
0.216 |
| 31 |
249253_at |
AT5G42060
|
[DEK, chromatin associated protein] |
0.143 |
-0.508 |
| 32 |
263772_at |
AT2G06255
|
ELF4-L3, ELF4-like 3 |
0.142 |
0.044 |
| 33 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
0.142 |
0.226 |
| 34 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.139 |
0.050 |
| 35 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.137 |
0.086 |
| 36 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.136 |
0.062 |
| 37 |
262605_at |
AT1G15170
|
[MATE efflux family protein] |
0.130 |
0.031 |
| 38 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
0.126 |
0.037 |
| 39 |
251576_at |
AT3G58200
|
[TRAF-like family protein] |
0.124 |
0.122 |
| 40 |
257952_at |
AT3G21770
|
[Peroxidase superfamily protein] |
0.120 |
0.027 |
| 41 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
0.116 |
0.030 |
| 42 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.116 |
0.700 |
| 43 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.115 |
0.218 |
| 44 |
254263_at |
AT4G23493
|
unknown |
0.115 |
-0.317 |
| 45 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.113 |
0.015 |
| 46 |
260472_at |
AT1G10990
|
unknown |
0.113 |
-0.051 |
| 47 |
256597_at |
AT3G28500
|
[60S acidic ribosomal protein family] |
0.113 |
0.090 |
| 48 |
261567_at |
AT1G33055
|
unknown |
0.112 |
0.007 |
| 49 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
0.112 |
0.123 |
| 50 |
249849_at |
AT5G23230
|
NIC2, nicotinamidase 2 |
0.112 |
0.071 |
| 51 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
0.111 |
-0.051 |
| 52 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
0.110 |
0.244 |
| 53 |
258604_at |
AT3G02980
|
MCC1, MEIOTIC CONTROL OF CROSSOVERS1 |
0.108 |
0.052 |
| 54 |
265025_at |
AT1G24575
|
unknown |
0.106 |
0.092 |
| 55 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.106 |
0.079 |
| 56 |
259325_at |
AT3G05320
|
[O-fucosyltransferase family protein] |
0.106 |
-0.286 |
| 57 |
266106_at |
AT2G45170
|
ATATG8E, AUTOPHAGY 8E, ATG8E, AUTOPHAGY 8E |
0.104 |
0.149 |
| 58 |
257191_at |
AT3G13175
|
unknown |
0.104 |
0.180 |
| 59 |
247167_at |
AT5G65850
|
[F-box and associated interaction domains-containing protein] |
0.104 |
0.134 |
| 60 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
0.103 |
0.136 |
| 61 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.101 |
-0.082 |
| 62 |
245034_at |
AT2G26390
|
[Serine protease inhibitor (SERPIN) family protein] |
0.101 |
0.439 |
| 63 |
253574_at |
AT4G31030
|
[Putative membrane lipoprotein] |
0.100 |
0.016 |
| 64 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
0.099 |
0.148 |
| 65 |
262756_at |
AT1G16370
|
ATOCT6, ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER 6, OCT6, organic cation/carnitine transporter 6 |
0.099 |
0.084 |
| 66 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.096 |
-0.214 |
| 67 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
0.096 |
0.006 |
| 68 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
0.096 |
0.199 |
| 69 |
258117_at |
AT3G14700
|
[SART-1 family] |
0.094 |
0.145 |
| 70 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
0.089 |
0.101 |
| 71 |
246888_at |
AT5G26270
|
unknown |
-0.825 |
-0.053 |
| 72 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.393 |
0.051 |
| 73 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
-0.389 |
-0.080 |
| 74 |
264780_at |
AT1G08720
|
ATEDR1, EDR1, ENHANCED DISEASE RESISTANCE 1 |
-0.249 |
-0.014 |
| 75 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-0.224 |
0.152 |
| 76 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
-0.213 |
0.008 |
| 77 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
-0.201 |
-0.113 |
| 78 |
260948_at |
AT1G06100
|
[Fatty acid desaturase family protein] |
-0.192 |
0.197 |
| 79 |
258893_at |
AT3G05660
|
AtRLP33, receptor like protein 33, RLP33, receptor like protein 33 |
-0.167 |
0.171 |
| 80 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
-0.152 |
-0.198 |
| 81 |
259953_at |
AT1G74810
|
BOR5, REQUIRES HIGH BORON 5 |
-0.149 |
-0.050 |
| 82 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.146 |
-0.115 |
| 83 |
250499_at |
AT5G09730
|
ATBX3, ATBXL3, BETA-XYLOSIDASE 3, BX3, BXL3, beta-xylosidase 3, XYL3 |
-0.146 |
0.193 |
| 84 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.145 |
-0.201 |
| 85 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
-0.144 |
-0.179 |
| 86 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.140 |
0.159 |
| 87 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.140 |
-0.109 |
| 88 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.135 |
-0.055 |
| 89 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.133 |
0.158 |
| 90 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.130 |
0.068 |
| 91 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
-0.128 |
-0.048 |
| 92 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
-0.126 |
0.226 |
| 93 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
-0.125 |
0.161 |
| 94 |
262891_at |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
-0.122 |
0.123 |
| 95 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
-0.121 |
0.064 |
| 96 |
254323_at |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
-0.119 |
0.280 |
| 97 |
266050_at |
AT2G40770
|
[zinc ion binding] |
-0.116 |
-0.153 |
| 98 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
-0.113 |
0.277 |
| 99 |
258392_at |
AT3G15400
|
ATA20, anther 20 |
-0.110 |
-0.046 |
| 100 |
260148_at |
AT1G52800
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.107 |
0.185 |
| 101 |
254759_at |
AT4G13180
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.105 |
-0.157 |
| 102 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
-0.103 |
0.051 |
| 103 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.102 |
0.029 |
| 104 |
251875_at |
AT3G54270
|
[sucrose-6F-phosphate phosphohydrolase family protein] |
-0.101 |
-0.096 |
| 105 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.101 |
-0.082 |
| 106 |
262450_at |
AT1G11320
|
unknown |
-0.099 |
0.067 |
| 107 |
254683_at |
AT4G13800
|
unknown |
-0.096 |
0.153 |
| 108 |
260776_at |
AT1G14580
|
[C2H2-like zinc finger protein] |
-0.096 |
0.159 |
| 109 |
245726_at |
AT1G73360
|
AtEDT1, ATHDG11, HOMEODOMAIN GLABROUS 11, EDT1, ENHANCED DROUGHT TOLERANCE 1, HDG11, homeodomain GLABROUS 11 |
-0.096 |
-0.098 |
| 110 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.094 |
0.135 |
| 111 |
259018_at |
AT3G07390
|
AIR12, Auxin-Induced in Root cultures 12 |
-0.094 |
-0.036 |
| 112 |
265539_at |
AT2G15830
|
unknown |
-0.092 |
0.516 |
| 113 |
263446_at |
AT2G31650
|
ATX1, homologue of trithorax, SDG27, SET DOMAIN PROTEIN 27 |
-0.091 |
-0.092 |
| 114 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.091 |
0.139 |
| 115 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.090 |
0.265 |
| 116 |
247351_at |
AT5G63790
|
ANAC102, NAC domain containing protein 102, NAC102, NAC domain containing protein 102 |
-0.089 |
0.008 |
| 117 |
267142_at |
AT2G38290
|
AMT2, ammonium transporter 2, AMT2;1, AMMONIUM TRANSPORTER 2;1, ATAMT2, ammonium transporter 2 |
-0.089 |
-0.057 |
| 118 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
-0.089 |
0.230 |
| 119 |
253997_at |
AT4G26090
|
RPS2, RESISTANT TO P. SYRINGAE 2 |
-0.088 |
0.138 |
| 120 |
266362_at |
AT2G32430
|
[Galactosyltransferase family protein] |
-0.088 |
0.138 |
| 121 |
260903_at |
AT1G02460
|
[Pectin lyase-like superfamily protein] |
-0.087 |
0.137 |
| 122 |
255259_at |
AT4G05020
|
NDB2, NAD(P)H dehydrogenase B2 |
-0.086 |
0.088 |
| 123 |
249336_at |
AT5G41070
|
DRB5, dsRNA-binding protein 5 |
-0.085 |
0.098 |
| 124 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.083 |
0.156 |
| 125 |
248706_at |
AT5G48530
|
unknown |
-0.082 |
0.151 |
| 126 |
245078_at |
AT2G23340
|
DEAR3, DREB and EAR motif protein 3 |
-0.082 |
-0.046 |
| 127 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
-0.081 |
0.233 |
| 128 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-0.080 |
0.076 |
| 129 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.080 |
0.100 |
| 130 |
263582_at |
AT2G17120
|
CL-1, CEBiP-like 1, LYM2, lysM domain GPI-anchored protein 2 precursor, LYP1, LysM-containing receptor protein 1 |
-0.080 |
-0.142 |
| 131 |
257276_at |
AT3G14460
|
[LRR and NB-ARC domains-containing disease resistance protein] |
-0.079 |
0.461 |
| 132 |
261799_at |
AT1G30473
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.079 |
0.186 |
| 133 |
263629_at |
AT2G04850
|
[Auxin-responsive family protein] |
-0.076 |
0.120 |
| 134 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.076 |
0.089 |
| 135 |
267572_at |
AT2G30660
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
-0.074 |
0.285 |
| 136 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.073 |
0.592 |
| 137 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.073 |
0.158 |
| 138 |
251051_at |
AT5G02460
|
[Dof-type zinc finger DNA-binding family protein] |
-0.072 |
0.161 |
| 139 |
265767_at |
AT2G48110
|
MED33B, REF4, REDUCED EPIDERMAL FLUORESCENCE 4 |
-0.072 |
0.038 |