|
probeID |
AGICode |
Annotation |
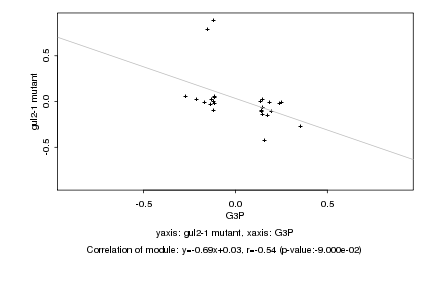
Log2 signal ratio
G3P |
Log2 signal ratio
gul2-1 mutant |
| 1 |
252661_at |
AT3G44450
|
unknown |
0.352 |
-0.270 |
| 2 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
0.250 |
0.000 |
| 3 |
259711_at |
AT1G77570
|
AtREN1, REN1, restricted to nucleolus 1 |
0.237 |
-0.014 |
| 4 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.196 |
-0.109 |
| 5 |
255887_at |
AT1G20370
|
[Pseudouridine synthase family protein] |
0.182 |
-0.004 |
| 6 |
245611_at |
AT4G14390
|
[Ankyrin repeat family protein] |
0.170 |
-0.151 |
| 7 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.158 |
-0.423 |
| 8 |
261048_at |
AT1G01420
|
UGT72B3, UDP-glucosyl transferase 72B3 |
0.147 |
-0.140 |
| 9 |
264392_at |
AT1G11900
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.147 |
0.023 |
| 10 |
263599_at |
AT2G01830
|
AHK4, ARABIDOPSIS HISTIDINE KINASE 4, ATCRE1, CRE1, CYTOKININ RESPONSE 1, WOL, WOODEN LEG, WOL1, WOODEN LEG 1 |
0.147 |
-0.061 |
| 11 |
249464_at |
AT5G39710
|
EMB2745, EMBRYO DEFECTIVE 2745 |
0.139 |
-0.101 |
| 12 |
254845_at |
AT4G11820
|
FKP1, FLAKY POLLEN 1, HMGS, HYDROXYMETHYLGLUTARYL-COA SYNTHASE, MVA1 |
0.139 |
-0.094 |
| 13 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
0.133 |
0.001 |
| 14 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.272 |
0.064 |
| 15 |
253717_at |
AT4G29440
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.216 |
0.025 |
| 16 |
250089_at |
AT5G17320
|
HDG9, homeodomain GLABROUS 9 |
-0.172 |
-0.005 |
| 17 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
-0.156 |
0.789 |
| 18 |
245962_at |
AT5G19700
|
[MATE efflux family protein] |
-0.139 |
-0.023 |
| 19 |
246209_at |
AT4G36870
|
BLH2, BEL1-like homeodomain 2, SAW1, SAWTOOTH 1 |
-0.132 |
0.032 |
| 20 |
263753_at |
AT2G21490
|
LEA, dehydrin LEA |
-0.123 |
0.896 |
| 21 |
262002_at |
AT1G64450
|
[Glycine-rich protein family] |
-0.122 |
-0.090 |
| 22 |
267176_at |
AT2G37730
|
unknown |
-0.120 |
0.006 |
| 23 |
253014_at |
AT4G37940
|
AGL21, AGAMOUS-like 21 |
-0.119 |
-0.020 |
| 24 |
263201_at |
AT1G05610
|
APS2, ADP-glucose pyrophosphorylase small subunit 2 |
-0.117 |
0.048 |
| 25 |
256238_at |
AT3G12400
|
ATELC, ELC |
-0.114 |
0.062 |