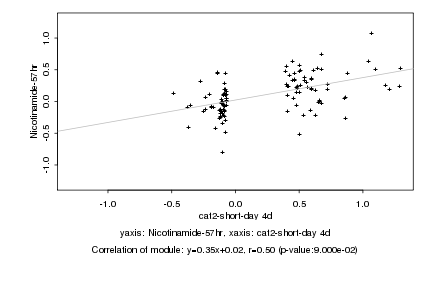
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
cat2-short-day 4d |
Log2 signal ratio
Nicotinamide-57hr |
| 1 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
1.292 |
0.523 |
| 2 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
1.282 |
0.242 |
| 3 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
1.205 |
0.197 |
| 4 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
1.171 |
0.262 |
| 5 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
1.095 |
0.507 |
| 6 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
1.066 |
1.080 |
| 7 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.041 |
0.639 |
| 8 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.878 |
0.449 |
| 9 |
251772_at |
AT3G55920
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.860 |
-0.264 |
| 10 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.856 |
0.067 |
| 11 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.855 |
0.063 |
| 12 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.716 |
0.279 |
| 13 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
0.715 |
0.191 |
| 14 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
0.673 |
0.743 |
| 15 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.671 |
0.511 |
| 16 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.670 |
-0.019 |
| 17 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.655 |
0.003 |
| 18 |
257670_at |
AT3G20340
|
unknown |
0.654 |
0.027 |
| 19 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.645 |
-0.013 |
| 20 |
260522_x_at |
AT2G41730
|
unknown |
0.640 |
0.536 |
| 21 |
250152_at |
AT5G15120
|
unknown |
0.626 |
-0.211 |
| 22 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.622 |
0.188 |
| 23 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.605 |
0.489 |
| 24 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.593 |
0.203 |
| 25 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.592 |
0.218 |
| 26 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.591 |
0.365 |
| 27 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.589 |
0.350 |
| 28 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.585 |
-0.140 |
| 29 |
248607_at |
AT5G49480
|
ATCP1, Ca2+-binding protein 1, CP1, Ca2+-binding protein 1 |
0.562 |
0.225 |
| 30 |
246584_at |
AT5G14730
|
unknown |
0.551 |
0.309 |
| 31 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.537 |
0.334 |
| 32 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.536 |
0.382 |
| 33 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.527 |
-0.216 |
| 34 |
259979_at |
AT1G76600
|
unknown |
0.509 |
0.268 |
| 35 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.509 |
0.499 |
| 36 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.501 |
0.578 |
| 37 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.501 |
-0.507 |
| 38 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.500 |
0.486 |
| 39 |
262935_at |
AT1G79410
|
AtOCT5, organic cation/carnitine transporter5, OCT5, organic cation/carnitine transporter5 |
0.498 |
0.157 |
| 40 |
263473_at |
AT2G31750
|
UGT74D1, UDP-glucosyl transferase 74D1 |
0.483 |
0.216 |
| 41 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.481 |
0.239 |
| 42 |
258805_at |
AT3G04010
|
[O-Glycosyl hydrolases family 17 protein] |
0.478 |
0.154 |
| 43 |
266296_at |
AT2G29420
|
ATGSTU7, glutathione S-transferase tau 7, GST25, GLUTATHIONE S-TRANSFERASE 25, GSTU7, glutathione S-transferase tau 7 |
0.474 |
0.233 |
| 44 |
262719_at |
AT1G43590
|
unknown |
0.472 |
-0.056 |
| 45 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.463 |
0.343 |
| 46 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.459 |
0.442 |
| 47 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.459 |
0.359 |
| 48 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
0.448 |
0.056 |
| 49 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.443 |
0.631 |
| 50 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
0.441 |
0.339 |
| 51 |
247287_at |
AT5G64230
|
unknown |
0.422 |
0.422 |
| 52 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.409 |
0.246 |
| 53 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.405 |
0.249 |
| 54 |
250910_at |
AT5G03720
|
AT-HSFA3, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A3, HSFA3, heat shock transcription factor A3 |
0.405 |
-0.145 |
| 55 |
245692_at |
AT5G04150
|
BHLH101 |
0.404 |
0.095 |
| 56 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.396 |
0.558 |
| 57 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.395 |
0.269 |
| 58 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
0.388 |
0.481 |
| 59 |
253174_at |
AT4G35090
|
CAT2, catalase 2 |
-0.491 |
0.142 |
| 60 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.383 |
-0.089 |
| 61 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-0.373 |
-0.398 |
| 62 |
248867_at |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
-0.359 |
-0.051 |
| 63 |
252309_at |
AT3G49340
|
[Cysteine proteinases superfamily protein] |
-0.278 |
0.317 |
| 64 |
255319_at |
AT4G04220
|
AtRLP46, receptor like protein 46, RLP46, receptor like protein 46 |
-0.258 |
-0.156 |
| 65 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
-0.238 |
0.075 |
| 66 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.235 |
-0.120 |
| 67 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.211 |
0.121 |
| 68 |
262811_at |
AT1G11700
|
unknown |
-0.192 |
-0.092 |
| 69 |
253687_at |
AT4G29520
|
[LOCATED IN: endoplasmic reticulum, plasma membrane] |
-0.191 |
-0.063 |
| 70 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
-0.178 |
-0.094 |
| 71 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.159 |
-0.419 |
| 72 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.146 |
0.465 |
| 73 |
248011_at |
AT5G56300
|
GAMT2, gibberellic acid methyltransferase 2 |
-0.142 |
0.449 |
| 74 |
264287_at |
AT1G61930
|
unknown |
-0.131 |
-0.129 |
| 75 |
265097_at |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
-0.126 |
-0.261 |
| 76 |
258546_at |
AT3G07060
|
emb1974, embryo defective 1974 |
-0.125 |
-0.123 |
| 77 |
256150_at |
AT1G55120
|
AtcwINV3, 6-fructan exohydrolase, ATFRUCT5, beta-fructofuranosidase 5, FRUCT5, beta-fructofuranosidase 5 |
-0.120 |
-0.186 |
| 78 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
-0.118 |
-0.250 |
| 79 |
256293_at |
AT1G69440
|
AGO7, ARGONAUTE7, ZIP, ZIPPY |
-0.118 |
-0.226 |
| 80 |
253654_at |
AT4G30060
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.116 |
0.044 |
| 81 |
258898_at |
AT3G05740
|
RECQI1, RECQ helicase l1 |
-0.116 |
-0.149 |
| 82 |
264696_at |
AT1G70230
|
AXY4, ALTERED XYLOGLUCAN 4, TBL27, TRICHOME BIREFRINGENCE-LIKE 27 |
-0.114 |
-0.023 |
| 83 |
247602_at |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
-0.113 |
0.000 |
| 84 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-0.106 |
-0.800 |
| 85 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
-0.105 |
-0.331 |
| 86 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.105 |
-0.184 |
| 87 |
254245_at |
AT4G23240
|
CRK16, cysteine-rich RLK (RECEPTOR-like protein kinase) 16 |
-0.102 |
0.003 |
| 88 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
-0.101 |
-0.047 |
| 89 |
262292_at |
AT1G27595
|
unknown |
-0.100 |
-0.046 |
| 90 |
260270_at |
AT1G63730
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.097 |
0.119 |
| 91 |
245565_at |
AT4G14605
|
MDA1, MTERF DEFECTIVE IN Arabidopsis1 |
-0.096 |
-0.111 |
| 92 |
259708_at |
AT1G77420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.094 |
-0.219 |
| 93 |
264666_at |
AT1G09680
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.094 |
0.102 |
| 94 |
265151_at |
AT1G51340
|
[MATE efflux family protein] |
-0.093 |
-0.065 |
| 95 |
250485_at |
AT5G09990
|
PROPEP5, elicitor peptide 5 precursor |
-0.092 |
0.128 |
| 96 |
265769_at |
AT2G48090
|
unknown |
-0.090 |
0.285 |
| 97 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.089 |
-0.231 |
| 98 |
262880_at |
AT1G64880
|
[Ribosomal protein S5 family protein] |
-0.088 |
-0.149 |
| 99 |
246429_at |
AT5G17450
|
HIPP21, heavy metal associated isoprenylated plant protein 21 |
-0.088 |
0.199 |
| 100 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
-0.088 |
-0.131 |
| 101 |
265978_at |
AT2G11140
|
[copia-like retrotransposon family, has a 1.0e-71 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-0.087 |
-0.062 |
| 102 |
251418_at |
AT3G60440
|
[Phosphoglycerate mutase family protein] |
-0.083 |
-0.296 |
| 103 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
-0.082 |
0.452 |
| 104 |
253175_at |
AT4G35050
|
MSI3, MULTICOPY SUPPRESSOR OF IRA1 3, NFC3, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 3 |
-0.081 |
0.113 |
| 105 |
262921_at |
AT1G79430
|
APL, ALTERED PHLOEM DEVELOPMENT, WDY, WOODY |
-0.081 |
0.097 |
| 106 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
-0.079 |
-0.476 |
| 107 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
-0.079 |
0.196 |
| 108 |
266110_at |
AT2G02080
|
AtIDD4, indeterminate(ID)-domain 4, IDD4, indeterminate(ID)-domain 4 |
-0.078 |
-0.058 |
| 109 |
251215_at |
AT3G62510
|
[protein disulfide isomerase-related] |
-0.077 |
0.014 |
| 110 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.076 |
0.056 |
| 111 |
250222_at |
AT5G14050
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.076 |
0.009 |
| 112 |
266917_at |
AT2G45830
|
DTA2, downstream target of AGL15 2 |
-0.075 |
0.114 |
| 113 |
248871_at |
AT5G46680
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.075 |
0.169 |
| 114 |
252247_at |
AT3G49740
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.074 |
0.021 |
| 115 |
261587_at |
AT1G01660
|
[RING/U-box superfamily protein] |
-0.073 |
0.063 |