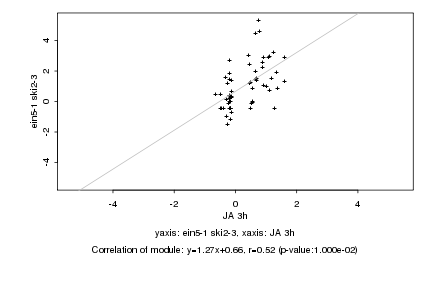
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
JA 3h |
Log2 signal ratio
ein5-1 ski2-3 |
| 1 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
1.596 |
2.894 |
| 2 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
1.591 |
1.377 |
| 3 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
1.344 |
0.890 |
| 4 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.310 |
1.964 |
| 5 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
1.252 |
-0.406 |
| 6 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
1.230 |
3.261 |
| 7 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
1.152 |
1.549 |
| 8 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.101 |
3.013 |
| 9 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
1.087 |
0.745 |
| 10 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.054 |
2.938 |
| 11 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
1.008 |
1.005 |
| 12 |
245209_at |
AT5G12340
|
unknown |
0.912 |
2.920 |
| 13 |
259384_at |
AT3G16450
|
JAL33, Jacalin-related lectin 33 |
0.902 |
1.078 |
| 14 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
0.881 |
2.264 |
| 15 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
0.853 |
2.569 |
| 16 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.761 |
4.640 |
| 17 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
0.737 |
5.379 |
| 18 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
0.672 |
1.435 |
| 19 |
256725_at |
AT2G34070
|
TBL37, TRICHOME BIREFRINGENCE-LIKE 37 |
0.662 |
1.544 |
| 20 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.650 |
4.512 |
| 21 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.643 |
2.019 |
| 22 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
0.625 |
1.460 |
| 23 |
257798_at |
AT3G15950
|
NAI2 |
0.557 |
0.031 |
| 24 |
266486_at |
AT2G47950
|
unknown |
0.540 |
0.890 |
| 25 |
263137_at |
AT1G78660
|
ATGGH1, gamma-glutamyl hydrolase 1, GGH1, gamma-glutamyl hydrolase 1 |
0.537 |
-0.030 |
| 26 |
262254_at |
AT1G53920
|
GLIP5, GDSL-motif lipase 5 |
0.512 |
-0.076 |
| 27 |
250252_at |
AT5G13750
|
ZIFL1, zinc induced facilitator-like 1 |
0.485 |
1.269 |
| 28 |
254064_at |
AT4G25410
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.482 |
-0.406 |
| 29 |
266456_at |
AT2G22770
|
NAI1 |
0.445 |
2.438 |
| 30 |
263032_at |
AT1G23850
|
unknown |
0.427 |
1.223 |
| 31 |
262171_at |
AT1G74950
|
JAZ2, JASMONATE-ZIM-DOMAIN PROTEIN 2, TIFY10B |
0.424 |
3.058 |
| 32 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.653 |
0.483 |
| 33 |
254013_at |
AT4G26050
|
PIRL8, plant intracellular ras group-related LRR 8 |
-0.520 |
-0.406 |
| 34 |
251491_at |
AT3G59480
|
[pfkB-like carbohydrate kinase family protein] |
-0.505 |
0.500 |
| 35 |
248931_at |
AT5G46040
|
[Major facilitator superfamily protein] |
-0.488 |
-0.406 |
| 36 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.413 |
-0.406 |
| 37 |
250398_at |
AT5G11000
|
unknown |
-0.327 |
1.593 |
| 38 |
249010_at |
AT5G44580
|
unknown |
-0.322 |
0.184 |
| 39 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.311 |
-0.978 |
| 40 |
254524_at |
AT4G20000
|
[VQ motif-containing protein] |
-0.287 |
1.197 |
| 41 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.280 |
-1.488 |
| 42 |
254118_at |
AT4G24790
|
[AAA-type ATPase family protein] |
-0.259 |
-0.112 |
| 43 |
256516_at |
AT1G66150
|
TMK1, transmembrane kinase 1 |
-0.217 |
0.459 |
| 44 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
-0.214 |
0.055 |
| 45 |
252987_at |
AT4G38390
|
RHS17, root hair specific 17 |
-0.213 |
-0.406 |
| 46 |
263194_at |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
-0.201 |
1.852 |
| 47 |
252836_at |
AT3G42070
|
unknown |
-0.200 |
0.246 |
| 48 |
265028_at |
AT1G24530
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.197 |
2.725 |
| 49 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.195 |
1.479 |
| 50 |
252176_at |
AT3G50720
|
[Protein kinase superfamily protein] |
-0.179 |
-0.406 |
| 51 |
249009_at |
AT5G44610
|
MAP18, microtubule-associated protein 18, PCAP2, PLASMA MEMBRANE ASSOCIATED CA2+-BINDING PROTEIN-2 |
-0.178 |
-0.406 |
| 52 |
245616_at |
AT4G14480
|
[Protein kinase superfamily protein] |
-0.176 |
-1.179 |
| 53 |
267634_at |
AT2G42100
|
[Actin-like ATPase superfamily protein] |
-0.175 |
-0.406 |
| 54 |
259769_at |
AT1G29400
|
AML5, MEI2-like protein 5, ML5, MEI2-like protein 5 |
-0.169 |
0.027 |
| 55 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
-0.168 |
0.278 |
| 56 |
251073_at |
AT5G01750
|
unknown |
-0.162 |
0.388 |
| 57 |
264181_at |
AT1G65350
|
UBQ13, ubiquitin 13 |
-0.160 |
0.268 |
| 58 |
253083_at |
AT4G36250
|
ALDH3F1, aldehyde dehydrogenase 3F1 |
-0.158 |
-0.719 |
| 59 |
251797_at |
AT3G55560
|
AGF2, AT-hook protein of GA feedback 2, AHL15, AT-hook motif nuclear-localized protein 15 |
-0.158 |
1.439 |
| 60 |
245237_at |
AT4G25520
|
SLK1, SEUSS-like 1 |
-0.156 |
0.363 |
| 61 |
247466_at |
AT5G62090
|
SLK2, SEUSS-like 2 |
-0.144 |
0.667 |