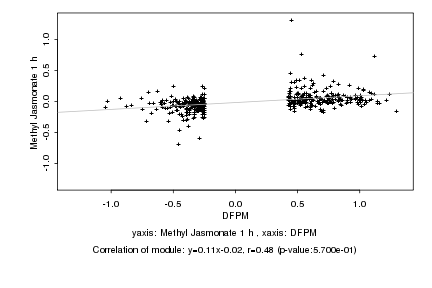
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
DFPM |
Log2 signal ratio
Methyl Jasmonate 1 h |
| 1 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
1.293 |
-0.147 |
| 2 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
1.238 |
0.117 |
| 3 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
1.211 |
0.018 |
| 4 |
249333_at |
AT5G40990
|
GLIP1, GDSL lipase 1 |
1.156 |
-0.027 |
| 5 |
265482_at |
AT2G15780
|
[Cupredoxin superfamily protein] |
1.138 |
-0.020 |
| 6 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
1.136 |
0.006 |
| 7 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
1.115 |
0.739 |
| 8 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
1.111 |
0.122 |
| 9 |
265658_at |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
1.093 |
0.130 |
| 10 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
1.087 |
0.044 |
| 11 |
249896_at |
AT5G22530
|
unknown |
1.076 |
0.023 |
| 12 |
260394_at |
AT1G74080
|
ATMYB122, MYB DOMAIN PROTEIN 122, MYB122, myb domain protein 122 |
1.076 |
0.158 |
| 13 |
251612_at |
AT3G57950
|
unknown |
1.054 |
-0.006 |
| 14 |
249438_at |
AT5G40010
|
AATP1, AAA-ATPase 1, ASD, ATPase-in-Seed-Development |
1.044 |
0.018 |
| 15 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
1.032 |
-0.046 |
| 16 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.030 |
0.180 |
| 17 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
1.025 |
0.196 |
| 18 |
254905_at |
AT4G11170
|
RMG1, Resistance Methylated Gene 1 |
1.020 |
0.026 |
| 19 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
1.014 |
0.072 |
| 20 |
252345_at |
AT3G48640
|
unknown |
1.011 |
0.048 |
| 21 |
262679_at |
AT1G75830
|
LCR67, low-molecular-weight cysteine-rich 67, PDF1.1, PLANT DEFENSIN 1.1 |
1.007 |
-0.075 |
| 22 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
1.005 |
-0.001 |
| 23 |
249001_at |
AT5G44990
|
[Glutathione S-transferase family protein] |
1.002 |
0.033 |
| 24 |
246406_at |
AT1G57650
|
[ATP binding] |
1.000 |
0.100 |
| 25 |
251971_at |
AT3G53160
|
UGT73C7, UDP-glucosyl transferase 73C7 |
0.997 |
0.030 |
| 26 |
266282_at |
AT2G29220
|
LecRK-III.1, L-type lectin receptor kinase III.1 |
0.991 |
0.046 |
| 27 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
0.989 |
0.212 |
| 28 |
261249_at |
AT1G05880
|
ARI12, ARIADNE 12, ATARI12, ARABIDOPSIS ARIADNE 12 |
0.989 |
-0.039 |
| 29 |
248319_at |
AT5G52710
|
[Copper transport protein family] |
0.979 |
-0.011 |
| 30 |
253044_at |
AT4G37290
|
unknown |
0.975 |
0.081 |
| 31 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.965 |
0.106 |
| 32 |
249495_at |
AT5G39100
|
GLP6, germin-like protein 6 |
0.964 |
0.048 |
| 33 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.961 |
0.061 |
| 34 |
247064_at |
AT5G66890
|
[Leucine-rich repeat (LRR) family protein] |
0.930 |
0.040 |
| 35 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.925 |
-0.015 |
| 36 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.924 |
0.067 |
| 37 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.923 |
-0.012 |
| 38 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.920 |
0.060 |
| 39 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.913 |
0.264 |
| 40 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
0.898 |
-0.008 |
| 41 |
251334_at |
AT3G61390
|
[RING/U-box superfamily protein] |
0.898 |
0.069 |
| 42 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
0.896 |
0.023 |
| 43 |
261466_at |
AT1G07690
|
unknown |
0.890 |
-0.028 |
| 44 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.882 |
0.096 |
| 45 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.862 |
0.103 |
| 46 |
254249_at |
AT4G23280
|
CRK20, cysteine-rich RLK (RECEPTOR-like protein kinase) 20 |
0.858 |
0.026 |
| 47 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.851 |
0.019 |
| 48 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
0.848 |
-0.062 |
| 49 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.844 |
0.043 |
| 50 |
262731_at |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
0.838 |
-0.048 |
| 51 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.831 |
0.065 |
| 52 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
0.827 |
0.279 |
| 53 |
259033_at |
AT3G09405
|
[Pectinacetylesterase family protein] |
0.827 |
0.076 |
| 54 |
248980_at |
AT5G45090
|
AtPP2-A7, phloem protein 2-A7, PP2-A7, phloem protein 2-A7 |
0.827 |
0.007 |
| 55 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.827 |
0.019 |
| 56 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.823 |
0.114 |
| 57 |
250415_at |
AT5G11210
|
ATGLR2.5, ARABIDOPSIS THALIANA GLU, GLR2.5, glutamate receptor 2.5 |
0.817 |
0.111 |
| 58 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.805 |
0.040 |
| 59 |
246984_at |
AT5G67310
|
CYP81G1, cytochrome P450, family 81, subfamily G, polypeptide 1 |
0.804 |
0.106 |
| 60 |
265705_at |
AT2G03410
|
[Mo25 family protein] |
0.795 |
-0.100 |
| 61 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.786 |
0.335 |
| 62 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.784 |
-0.002 |
| 63 |
263854_at |
AT2G04430
|
atnudt5, nudix hydrolase homolog 5, NUDT5, nudix hydrolase homolog 5 |
0.783 |
0.104 |
| 64 |
258203_at |
AT3G13950
|
unknown |
0.782 |
-0.009 |
| 65 |
252421_at |
AT3G47540
|
[Chitinase family protein] |
0.778 |
0.086 |
| 66 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.776 |
0.068 |
| 67 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.776 |
0.040 |
| 68 |
246370_at |
AT1G51920
|
unknown |
0.776 |
-0.016 |
| 69 |
247145_at |
AT5G65600
|
LecRK-IX.2, L-type lectin receptor kinase IX.2 |
0.772 |
0.137 |
| 70 |
262085_at |
AT1G56060
|
unknown |
0.769 |
0.145 |
| 71 |
254922_at |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
0.768 |
-0.010 |
| 72 |
249004_at |
AT5G44570
|
unknown |
0.762 |
-0.008 |
| 73 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.758 |
0.251 |
| 74 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
0.752 |
0.021 |
| 75 |
247913_at |
AT5G57510
|
unknown |
0.751 |
0.059 |
| 76 |
259921_at |
AT1G72540
|
[Protein kinase superfamily protein] |
0.748 |
0.064 |
| 77 |
248971_at |
AT5G45000
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.744 |
-0.019 |
| 78 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.742 |
0.090 |
| 79 |
245363_at |
AT4G15120
|
[VQ motif-containing protein] |
0.739 |
0.113 |
| 80 |
267391_at |
AT2G44480
|
BGLU17, beta glucosidase 17 |
0.734 |
-0.004 |
| 81 |
251456_at |
AT3G60120
|
BGLU27, beta glucosidase 27 |
0.734 |
-0.030 |
| 82 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.733 |
0.026 |
| 83 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
0.728 |
0.090 |
| 84 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
0.728 |
-0.015 |
| 85 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
0.725 |
0.223 |
| 86 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
0.714 |
0.018 |
| 87 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.711 |
0.001 |
| 88 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.708 |
0.175 |
| 89 |
254292_at |
AT4G23030
|
[MATE efflux family protein] |
0.706 |
-0.132 |
| 90 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
0.705 |
-0.166 |
| 91 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
0.704 |
0.431 |
| 92 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.700 |
-0.009 |
| 93 |
261394_at |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
0.691 |
0.044 |
| 94 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
0.690 |
0.045 |
| 95 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.690 |
-0.153 |
| 96 |
254014_at |
AT4G26120
|
[Ankyrin repeat family protein / BTB/POZ domain-containing protein] |
0.684 |
-0.121 |
| 97 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.683 |
0.061 |
| 98 |
256176_at |
AT1G51640
|
ATEXO70G2, exocyst subunit exo70 family protein G2, EXO70G2, exocyst subunit exo70 family protein G2 |
0.679 |
-0.048 |
| 99 |
260648_at |
AT1G08050
|
[Zinc finger (C3HC4-type RING finger) family protein] |
0.679 |
0.061 |
| 100 |
266052_at |
AT2G40740
|
ATWRKY55, WRKY DNA-BINDING PROTEIN 55, WRKY55, WRKY DNA-binding protein 55 |
0.677 |
0.049 |
| 101 |
247618_at |
AT5G60280
|
LecRK-I.8, L-type lectin receptor kinase I.8 |
0.672 |
0.051 |
| 102 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.672 |
-0.001 |
| 103 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.671 |
0.029 |
| 104 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.663 |
-0.029 |
| 105 |
257978_at |
AT3G20860
|
ATNEK5, NIMA-related kinase 5, NEK5, NIMA-related kinase 5 |
0.657 |
0.013 |
| 106 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.651 |
0.170 |
| 107 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.648 |
0.075 |
| 108 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
0.646 |
0.086 |
| 109 |
249255_at |
AT5G41610
|
ATCHX18, ARABIDOPSIS THALIANA CATION/H+ EXCHANGER 18, CHX18, cation/H+ exchanger 18 |
0.638 |
-0.078 |
| 110 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.630 |
0.157 |
| 111 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.626 |
0.292 |
| 112 |
248536_at |
AT5G50140
|
[Ankyrin repeat family protein] |
0.625 |
0.030 |
| 113 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.618 |
0.046 |
| 114 |
249562_at |
AT5G38350
|
[Disease resistance protein (NBS-LRR class) family] |
0.615 |
-0.059 |
| 115 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
0.614 |
0.272 |
| 116 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
0.611 |
0.001 |
| 117 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.609 |
-0.056 |
| 118 |
245662_at |
AT1G28190
|
unknown |
0.608 |
0.342 |
| 119 |
249096_at |
AT5G43910
|
[pfkB-like carbohydrate kinase family protein] |
0.603 |
-0.119 |
| 120 |
250796_at |
AT5G05300
|
unknown |
0.603 |
0.065 |
| 121 |
258577_at |
AT3G04220
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.600 |
0.112 |
| 122 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
0.600 |
-0.082 |
| 123 |
250267_at |
AT5G12930
|
unknown |
0.600 |
-0.029 |
| 124 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.600 |
0.067 |
| 125 |
254408_at |
AT4G21390
|
B120 |
0.599 |
0.222 |
| 126 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.597 |
0.059 |
| 127 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.594 |
0.109 |
| 128 |
254289_at |
AT4G22980
|
unknown |
0.592 |
0.044 |
| 129 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
0.580 |
0.111 |
| 130 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.575 |
-0.080 |
| 131 |
249889_at |
AT5G22540
|
unknown |
0.571 |
0.140 |
| 132 |
262228_at |
AT1G68690
|
AtPERK9, proline-rich extensin-like receptor kinase 9, PERK9, proline-rich extensin-like receptor kinase 9 |
0.570 |
0.040 |
| 133 |
249055_at |
AT5G44460
|
CML43, calmodulin like 43 |
0.568 |
0.066 |
| 134 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.566 |
0.075 |
| 135 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.564 |
-0.002 |
| 136 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.563 |
0.020 |
| 137 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.562 |
-0.085 |
| 138 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.558 |
-0.024 |
| 139 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
0.557 |
0.020 |
| 140 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.554 |
0.136 |
| 141 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
0.553 |
0.258 |
| 142 |
252684_at |
AT3G44400
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.553 |
0.029 |
| 143 |
251769_at |
AT3G55950
|
ATCRR3, CCR3, CRINKLY4 related 3 |
0.551 |
0.378 |
| 144 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.550 |
0.000 |
| 145 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.546 |
0.107 |
| 146 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
0.545 |
-0.013 |
| 147 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.540 |
-0.011 |
| 148 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.539 |
0.034 |
| 149 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.538 |
0.037 |
| 150 |
253963_at |
AT4G26470
|
[Calcium-binding EF-hand family protein] |
0.537 |
0.071 |
| 151 |
250695_at |
AT5G06740
|
LecRK-S.5, L-type lectin receptor kinase S.5 |
0.537 |
0.005 |
| 152 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.535 |
0.105 |
| 153 |
266711_at |
AT2G46740
|
AtGulLO5, GulLO5, L -gulono-1,4-lactone ( L -GulL) oxidase 5 |
0.534 |
0.069 |
| 154 |
255074_at |
AT4G09100
|
[RING/U-box superfamily protein] |
0.533 |
0.034 |
| 155 |
249767_at |
AT5G24090
|
ATCHIA, chitinase A, CHIA, chitinase A |
0.531 |
0.088 |
| 156 |
245076_at |
AT2G23170
|
GH3.3 |
0.530 |
0.212 |
| 157 |
247922_at |
AT5G57500
|
[Galactosyltransferase family protein] |
0.527 |
-0.025 |
| 158 |
252862_at |
AT4G39830
|
[Cupredoxin superfamily protein] |
0.527 |
0.064 |
| 159 |
250149_at |
AT5G14700
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.526 |
0.774 |
| 160 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.520 |
-0.028 |
| 161 |
261481_at |
AT1G14260
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.519 |
-0.045 |
| 162 |
245197_at |
AT1G67800
|
[Copine (Calcium-dependent phospholipid-binding protein) family] |
0.516 |
-0.050 |
| 163 |
245454_at |
AT4G16920
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.515 |
0.031 |
| 164 |
261161_at |
AT1G34420
|
[leucine-rich repeat transmembrane protein kinase family protein] |
0.515 |
0.004 |
| 165 |
262942_at |
AT1G79450
|
ALIS5, ALA-interacting subunit 5 |
0.512 |
0.014 |
| 166 |
266780_at |
AT2G29110
|
ATGLR2.8, glutamate receptor 2.8, GLR2.8, GLR2.8, glutamate receptor 2.8 |
0.510 |
0.040 |
| 167 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
0.508 |
0.348 |
| 168 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
0.508 |
-0.022 |
| 169 |
259489_at |
AT1G15790
|
unknown |
0.506 |
0.117 |
| 170 |
247594_at |
AT5G60800
|
[Heavy metal transport/detoxification superfamily protein ] |
0.506 |
0.135 |
| 171 |
255599_at |
AT4G01010
|
ATCNGC13, CYCLIC NUCLEOTIDE-GATED CHANNEL 13, CNGC13, cyclic nucleotide-gated channel 13 |
0.506 |
-0.010 |
| 172 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.500 |
0.001 |
| 173 |
262542_at |
AT1G34180
|
anac016, NAC domain containing protein 16, NAC016, NAC domain containing protein 16 |
0.499 |
0.019 |
| 174 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.498 |
0.075 |
| 175 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.496 |
-0.018 |
| 176 |
258002_at |
AT3G28930
|
AIG2, AVRRPT2-INDUCED GENE 2 |
0.493 |
0.138 |
| 177 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.490 |
0.115 |
| 178 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.490 |
0.235 |
| 179 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.486 |
0.340 |
| 180 |
248416_at |
AT5G51630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.483 |
-0.019 |
| 181 |
245566_at |
AT4G14610
|
[pseudogene] |
0.477 |
0.021 |
| 182 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.475 |
0.080 |
| 183 |
254673_at |
AT4G18430
|
AtRABA1e, RAB GTPase homolog A1E, RABA1e, RAB GTPase homolog A1E |
0.474 |
-0.032 |
| 184 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.472 |
-0.160 |
| 185 |
260754_at |
AT1G49000
|
unknown |
0.470 |
-0.078 |
| 186 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.470 |
0.021 |
| 187 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.469 |
-0.106 |
| 188 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.469 |
0.317 |
| 189 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.468 |
-0.024 |
| 190 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.467 |
-0.075 |
| 191 |
254575_at |
AT4G19460
|
[UDP-Glycosyltransferase superfamily protein] |
0.464 |
-0.014 |
| 192 |
246841_at |
AT5G26700
|
[RmlC-like cupins superfamily protein] |
0.452 |
-0.001 |
| 193 |
259737_at |
AT1G64400
|
LACS3, long-chain acyl-CoA synthetase 3 |
0.448 |
0.024 |
| 194 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.447 |
0.319 |
| 195 |
255844_at |
AT2G33580
|
LYK5, LysM-containing receptor-like kinase 5 |
0.446 |
-0.019 |
| 196 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.445 |
1.325 |
| 197 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.443 |
0.172 |
| 198 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.442 |
-0.072 |
| 199 |
254847_at |
AT4G11850
|
MEE54, maternal effect embryo arrest 54, PLDGAMMA1, phospholipase D gamma 1 |
0.442 |
0.087 |
| 200 |
249314_at |
AT5G41180
|
[leucine-rich repeat transmembrane protein kinase family protein] |
0.441 |
-0.021 |
| 201 |
246584_at |
AT5G14730
|
unknown |
0.438 |
0.455 |
| 202 |
248381_at |
AT5G51830
|
[pfkB-like carbohydrate kinase family protein] |
0.437 |
0.035 |
| 203 |
253012_at |
AT4G37900
|
unknown |
0.436 |
-0.115 |
| 204 |
251106_at |
AT5G01500
|
TAAC, thylakoid ATP/ADP carrier |
0.435 |
0.087 |
| 205 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.435 |
0.222 |
| 206 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.434 |
0.074 |
| 207 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.432 |
0.169 |
| 208 |
259018_at |
AT3G07390
|
AIR12, Auxin-Induced in Root cultures 12 |
0.430 |
0.030 |
| 209 |
256306_at |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
0.429 |
-0.024 |
| 210 |
260179_at |
AT1G70690
|
HWI1, HOPW1-1-INDUCED GENE1, PDLP5, PLASMODESMATA-LOCATED PROTEIN 5 |
0.427 |
0.144 |
| 211 |
259019_at |
AT3G07370
|
ATCHIP, CARBOXYL TERMINUS OF HSC70-INTERACTING PROTEIN, CHIP, carboxyl terminus of HSC70-interacting protein |
0.425 |
-0.061 |
| 212 |
257830_at |
AT3G26690
|
ATNUDT13, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 13, ATNUDX13, nudix hydrolase homolog 13, NUDX13, nudix hydrolase homolog 13 |
0.425 |
0.086 |
| 213 |
255319_at |
AT4G04220
|
AtRLP46, receptor like protein 46, RLP46, receptor like protein 46 |
0.424 |
0.069 |
| 214 |
253124_at |
AT4G36030
|
ARO3, armadillo repeat only 3 |
0.423 |
0.021 |
| 215 |
258108_at |
AT3G23570
|
[alpha/beta-Hydrolases superfamily protein] |
0.422 |
0.035 |
| 216 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-1.049 |
-0.091 |
| 217 |
265400_at |
AT2G10940
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.034 |
0.003 |
| 218 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.930 |
0.052 |
| 219 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
-0.879 |
-0.075 |
| 220 |
267459_at |
AT2G33850
|
unknown |
-0.844 |
-0.064 |
| 221 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-0.760 |
0.054 |
| 222 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.748 |
-0.114 |
| 223 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.717 |
-0.318 |
| 224 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.703 |
0.160 |
| 225 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.699 |
-0.026 |
| 226 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.678 |
-0.187 |
| 227 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
-0.662 |
-0.016 |
| 228 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
-0.640 |
-0.120 |
| 229 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.628 |
0.172 |
| 230 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.606 |
-0.004 |
| 231 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
-0.603 |
-0.003 |
| 232 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.599 |
-0.042 |
| 233 |
252272_at |
AT3G49670
|
BAM2, BARELY ANY MERISTEM 2 |
-0.593 |
-0.088 |
| 234 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-0.588 |
0.022 |
| 235 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.576 |
-0.029 |
| 236 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.564 |
-0.097 |
| 237 |
259660_at |
AT1G55260
|
LTPG6, glycosylphosphatidylinositol-anchored lipid protein transfer 6 |
-0.558 |
0.021 |
| 238 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.551 |
-0.098 |
| 239 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.544 |
-0.312 |
| 240 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.532 |
-0.180 |
| 241 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
-0.530 |
-0.088 |
| 242 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-0.525 |
-0.010 |
| 243 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
-0.521 |
0.093 |
| 244 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.512 |
-0.168 |
| 245 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.509 |
-0.064 |
| 246 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.506 |
0.002 |
| 247 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-0.506 |
0.243 |
| 248 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.504 |
-0.053 |
| 249 |
260883_at |
AT1G29270
|
unknown |
-0.489 |
0.037 |
| 250 |
252215_at |
AT3G50240
|
KICP-02 |
-0.486 |
-0.074 |
| 251 |
251395_at |
AT2G45470
|
AGP8, ARABINOGALACTAN PROTEIN 8, FLA8, FASCICLIN-like arabinogalactan protein 8 |
-0.482 |
0.012 |
| 252 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.481 |
-0.129 |
| 253 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.478 |
-0.066 |
| 254 |
253270_at |
AT4G34160
|
CYCD3, CYCD3;1, CYCLIN D3;1 |
-0.474 |
-0.134 |
| 255 |
252971_at |
AT4G38770
|
ATPRP4, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 4, PRP4, proline-rich protein 4 |
-0.474 |
-0.010 |
| 256 |
247474_at |
AT5G62280
|
unknown |
-0.465 |
-0.685 |
| 257 |
262260_at |
AT1G70850
|
MLP34, MLP-like protein 34 |
-0.459 |
-0.024 |
| 258 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.458 |
-0.203 |
| 259 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
-0.456 |
0.012 |
| 260 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
-0.454 |
-0.032 |
| 261 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.454 |
-0.065 |
| 262 |
248028_at |
AT5G55620
|
unknown |
-0.451 |
-0.456 |
| 263 |
262728_at |
AT1G75820
|
ATCLV1, CLV1, CLAVATA 1, FAS3, FASCIATA 3, FLO5, FLOWER DEVELOPMENT 5 |
-0.443 |
-0.081 |
| 264 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.442 |
-0.206 |
| 265 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
-0.433 |
-0.030 |
| 266 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.431 |
-0.053 |
| 267 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.430 |
-0.226 |
| 268 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
-0.428 |
-0.023 |
| 269 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.426 |
-0.298 |
| 270 |
250366_at |
AT5G11420
|
unknown |
-0.424 |
-0.069 |
| 271 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.423 |
0.058 |
| 272 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.422 |
-0.306 |
| 273 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.418 |
-0.043 |
| 274 |
248427_at |
AT5G51750
|
ATSBT1.3, subtilase 1.3, SBT1.3, subtilase 1.3 |
-0.414 |
-0.032 |
| 275 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
-0.410 |
-0.009 |
| 276 |
258480_at |
AT3G02640
|
unknown |
-0.406 |
-0.015 |
| 277 |
261203_at |
AT1G12845
|
unknown |
-0.405 |
0.019 |
| 278 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.400 |
-0.215 |
| 279 |
259763_at |
AT1G77630
|
LYM3, lysin-motif (LysM) domain protein 3, LYP3, LysM-containing receptor protein 3 |
-0.400 |
-0.124 |
| 280 |
262312_at |
AT1G70830
|
MLP28, MLP-like protein 28 |
-0.399 |
0.015 |
| 281 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.398 |
-0.014 |
| 282 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.397 |
-0.301 |
| 283 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.394 |
-0.166 |
| 284 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.394 |
-0.111 |
| 285 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
-0.393 |
0.068 |
| 286 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.389 |
-0.285 |
| 287 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.387 |
-0.102 |
| 288 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
-0.387 |
-0.174 |
| 289 |
254982_at |
AT4G10470
|
unknown |
-0.385 |
-0.061 |
| 290 |
258376_at |
AT3G17680
|
[Kinase interacting (KIP1-like) family protein] |
-0.381 |
0.061 |
| 291 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.379 |
-0.397 |
| 292 |
246505_at |
AT5G16250
|
unknown |
-0.377 |
-0.039 |
| 293 |
255957_at |
AT1G22160
|
unknown |
-0.376 |
-0.139 |
| 294 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
-0.375 |
-0.198 |
| 295 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.373 |
0.007 |
| 296 |
262230_at |
AT1G68560
|
ATXYL1, ALPHA-XYLOSIDASE 1, AXY3, altered xyloglucan 3, TRG1, thermoinhibition resistant germination 1, XYL1, alpha-xylosidase 1 |
-0.371 |
-0.005 |
| 297 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-0.371 |
-0.029 |
| 298 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
-0.371 |
-0.122 |
| 299 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-0.370 |
-0.017 |
| 300 |
247182_at |
AT5G65410
|
ATHB25, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 25, HB25, homeobox protein 25, ZFHD2, ZINC FINGER HOMEODOMAIN 2, ZHD1, ZINC FINGER HOMEODOMAIN 1 |
-0.370 |
-0.163 |
| 301 |
254649_at |
AT4G18570
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.366 |
-0.038 |
| 302 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.365 |
-0.099 |
| 303 |
264377_at |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
-0.361 |
0.003 |
| 304 |
263209_at |
AT1G10522
|
PRIN2, PLASTID REDOX INSENSITIVE 2 |
-0.359 |
-0.028 |
| 305 |
248772_at |
AT5G47800
|
[Phototropic-responsive NPH3 family protein] |
-0.358 |
-0.033 |
| 306 |
249718_at |
AT5G35740
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.356 |
-0.009 |
| 307 |
250690_at |
AT5G06530
|
ABCG22, ATP-binding cassette G22, AtABCG22, Arabidopsis thaliana ATP-binding cassette G22 |
-0.356 |
-0.095 |
| 308 |
247826_at |
AT5G58480
|
[O-Glycosyl hydrolases family 17 protein] |
-0.355 |
-0.047 |
| 309 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
-0.353 |
-0.020 |
| 310 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
-0.352 |
-0.048 |
| 311 |
254466_at |
AT4G20430
|
[Subtilase family protein] |
-0.352 |
-0.043 |
| 312 |
257236_at |
AT3G15095
|
HCF243, high chlorophyll fluorescence 243 |
-0.349 |
-0.095 |
| 313 |
260902_at |
AT1G21440
|
[Phosphoenolpyruvate carboxylase family protein] |
-0.347 |
0.039 |
| 314 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.345 |
-0.059 |
| 315 |
245130_at |
AT2G45340
|
[Leucine-rich repeat protein kinase family protein] |
-0.344 |
0.019 |
| 316 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.344 |
-0.006 |
| 317 |
247450_at |
AT5G62350
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.341 |
0.093 |
| 318 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
-0.341 |
0.075 |
| 319 |
246231_at |
AT4G37080
|
unknown |
-0.338 |
-0.259 |
| 320 |
254789_at |
AT4G12880
|
AtENODL19, ENODL19, early nodulin-like protein 19 |
-0.337 |
0.014 |
| 321 |
249807_at |
AT5G23870
|
[Pectinacetylesterase family protein] |
-0.337 |
-0.250 |
| 322 |
248420_at |
AT5G51560
|
[Leucine-rich repeat protein kinase family protein] |
-0.337 |
-0.196 |
| 323 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.336 |
-0.061 |
| 324 |
247288_at |
AT5G64330
|
JK218, NPH3, NON-PHOTOTROPIC HYPOCOTYL 3, RPT3, ROOT PHOTOTROPISM 3 |
-0.336 |
-0.172 |
| 325 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.336 |
-0.202 |
| 326 |
256676_at |
AT3G52180
|
ATPTPKIS1, ATSEX4, DSP4, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 4, SEX4, STARCH-EXCESS 4 |
-0.335 |
0.033 |
| 327 |
266598_at |
AT2G46110
|
KPHMT1, ketopantoate hydroxymethyltransferase 1, PANB1 |
-0.333 |
-0.060 |
| 328 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.333 |
-0.061 |
| 329 |
264497_at |
AT1G30840
|
ATPUP4, purine permease 4, PUP4, purine permease 4 |
-0.331 |
-0.127 |
| 330 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.330 |
-0.165 |
| 331 |
247037_at |
AT5G67070
|
RALFL34, ralf-like 34 |
-0.330 |
0.073 |
| 332 |
262263_at |
AT1G70940
|
ATPIN3, ARABIDOPSIS PIN-FORMED 3, PIN3, PIN-FORMED 3 |
-0.330 |
-0.083 |
| 333 |
264315_at |
AT1G70370
|
PG2, polygalacturonase 2 |
-0.327 |
-0.024 |
| 334 |
247541_at |
AT5G61660
|
[glycine-rich protein] |
-0.327 |
-0.025 |
| 335 |
251174_at |
AT3G63200
|
PLA IIIB, PLP9, PATATIN-like protein 9 |
-0.327 |
-0.056 |
| 336 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.327 |
-0.054 |
| 337 |
252954_at |
AT4G38660
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.324 |
-0.044 |
| 338 |
264371_at |
AT1G12090
|
ELP, extensin-like protein |
-0.321 |
0.028 |
| 339 |
245845_at |
AT1G26150
|
AtPERK10, proline-rich extensin-like receptor kinase 10, PERK10, proline-rich extensin-like receptor kinase 10 |
-0.319 |
-0.054 |
| 340 |
253749_at |
AT4G29080
|
IAA27, indole-3-acetic acid inducible 27, PAP2, phytochrome-associated protein 2 |
-0.318 |
-0.005 |
| 341 |
245296_at |
AT4G16370
|
oligopeptide transporter |
-0.316 |
-0.130 |
| 342 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.315 |
-0.063 |
| 343 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
-0.313 |
-0.024 |
| 344 |
256960_at |
AT3G13510
|
unknown |
-0.312 |
-0.108 |
| 345 |
255460_at |
AT4G02800
|
unknown |
-0.312 |
-0.060 |
| 346 |
247340_at |
AT5G63700
|
[zinc ion binding] |
-0.311 |
-0.115 |
| 347 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.311 |
-0.043 |
| 348 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.309 |
-0.010 |
| 349 |
251670_at |
AT3G57190
|
PrfB3, peptide chain release factor 3 |
-0.308 |
-0.146 |
| 350 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.308 |
-0.061 |
| 351 |
264900_at |
AT1G23080
|
ATPIN7, ARABIDOPSIS PIN-FORMED 7, PIN7, PIN-FORMED 7 |
-0.308 |
-0.129 |
| 352 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.306 |
-0.005 |
| 353 |
256784_at |
AT3G13674
|
unknown |
-0.306 |
-0.101 |
| 354 |
264078_at |
AT2G28470
|
BGAL8, beta-galactosidase 8 |
-0.304 |
-0.096 |
| 355 |
254133_at |
AT4G24810
|
[Protein kinase superfamily protein] |
-0.304 |
-0.067 |
| 356 |
260107_at |
AT1G66430
|
[pfkB-like carbohydrate kinase family protein] |
-0.303 |
-0.010 |
| 357 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.302 |
-0.156 |
| 358 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.300 |
-0.056 |
| 359 |
263902_at |
AT2G36230
|
APG10, ALBINO AND PALE GREEN 10, HISN3 |
-0.297 |
-0.072 |
| 360 |
257912_at |
AT3G25500
|
AFH1, formin homology 1, AHF1, ATFH1, ARABIDOPSIS THALIANA FORMIN HOMOLOGY 1, FH1, FORMIN HOMOLOGY 1 |
-0.297 |
-0.043 |
| 361 |
253218_at |
AT4G34980
|
SLP2, subtilisin-like serine protease 2 |
-0.294 |
-0.101 |
| 362 |
262162_at |
AT1G78020
|
unknown |
-0.290 |
-0.139 |
| 363 |
249347_at |
AT5G40830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.290 |
0.043 |
| 364 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.290 |
-0.594 |
| 365 |
251374_at |
AT3G60390
|
HAT3, homeobox-leucine zipper protein 3 |
-0.289 |
-0.125 |
| 366 |
251083_at |
AT5G01590
|
TIC56, TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 56 |
-0.289 |
-0.043 |
| 367 |
259000_at |
AT3G01860
|
unknown |
-0.288 |
-0.122 |
| 368 |
258222_at |
AT3G15680
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-0.288 |
-0.107 |
| 369 |
267465_at |
AT2G19170
|
SLP3, subtilisin-like serine protease 3 |
-0.287 |
-0.007 |
| 370 |
246919_at |
AT5G25460
|
DGR2, DUF642 L-GalL responsive gene 2 |
-0.284 |
-0.026 |
| 371 |
253227_at |
AT4G35030
|
[Protein kinase superfamily protein] |
-0.283 |
-0.131 |
| 372 |
255104_at |
AT4G08685
|
SAH7 |
-0.282 |
-0.016 |
| 373 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
-0.282 |
0.071 |
| 374 |
249826_at |
AT5G23310
|
FSD3, Fe superoxide dismutase 3 |
-0.280 |
-0.054 |
| 375 |
257510_at |
AT1G55360
|
unknown |
-0.280 |
-0.078 |
| 376 |
247586_at |
AT5G60660
|
PIP2;4, plasma membrane intrinsic protein 2;4, PIP2F |
-0.279 |
0.019 |
| 377 |
248186_at |
AT5G53880
|
unknown |
-0.278 |
-0.030 |
| 378 |
253875_at |
AT4G27520
|
AtENODL2, ENODL2, early nodulin-like protein 2 |
-0.277 |
-0.033 |
| 379 |
251278_at |
AT3G61780
|
emb1703, embryo defective 1703 |
-0.276 |
-0.016 |
| 380 |
248277_at |
AT5G52860
|
ABCG8, ATP-binding cassette G8 |
-0.276 |
0.074 |
| 381 |
261301_at |
AT1G48570
|
[zinc finger (Ran-binding) family protein] |
-0.274 |
-0.012 |
| 382 |
262388_at |
AT1G49320
|
unknown |
-0.273 |
-0.121 |
| 383 |
257204_at |
AT3G23805
|
RALFL24, ralf-like 24 |
-0.273 |
-0.052 |
| 384 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.272 |
-0.200 |
| 385 |
258424_at |
AT3G16750
|
unknown |
-0.272 |
-0.051 |
| 386 |
246475_at |
AT5G16720
|
MyoB3, myosin binding protein 3 |
-0.272 |
-0.039 |
| 387 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.272 |
-0.040 |
| 388 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-0.271 |
-0.008 |
| 389 |
245186_at |
AT1G67710
|
ARR11, response regulator 11 |
-0.271 |
-0.086 |
| 390 |
261055_at |
AT1G01300
|
[Eukaryotic aspartyl protease family protein] |
-0.271 |
0.024 |
| 391 |
245859_at |
AT5G28290
|
ATNEK3, NIMA-related kinase 3, NEK3, NIMA-related kinase 3 |
-0.270 |
-0.029 |
| 392 |
260347_at |
AT1G69420
|
[DHHC-type zinc finger family protein] |
-0.270 |
-0.081 |
| 393 |
261248_at |
AT1G20030
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.269 |
0.129 |
| 394 |
256626_at |
AT3G20015
|
[Eukaryotic aspartyl protease family protein] |
-0.269 |
-0.154 |
| 395 |
262738_at |
AT1G28530
|
unknown |
-0.268 |
0.030 |
| 396 |
258912_at |
AT3G06460
|
[GNS1/SUR4 membrane protein family] |
-0.268 |
0.027 |
| 397 |
258080_at |
AT3G25930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.268 |
-0.247 |
| 398 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.267 |
-0.068 |
| 399 |
247191_at |
AT5G65310
|
ATHB-5, ATHB5, homeobox protein 5, HB5, homeobox protein 5 |
-0.267 |
-0.064 |
| 400 |
266526_at |
AT2G16980
|
[Major facilitator superfamily protein] |
-0.266 |
0.244 |
| 401 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.266 |
-0.069 |
| 402 |
251119_at |
AT3G63510
|
[FMN-linked oxidoreductases superfamily protein] |
-0.266 |
-0.051 |
| 403 |
259858_at |
AT1G68400
|
[leucine-rich repeat transmembrane protein kinase family protein] |
-0.266 |
-0.070 |
| 404 |
252033_at |
AT3G51950
|
[Zinc finger (CCCH-type) family protein / RNA recognition motif (RRM)-containing protein] |
-0.265 |
-0.036 |
| 405 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.265 |
0.004 |
| 406 |
266022_at |
AT2G05920
|
[Subtilase family protein] |
-0.265 |
-0.017 |
| 407 |
251287_at |
AT3G61820
|
[Eukaryotic aspartyl protease family protein] |
-0.263 |
0.010 |
| 408 |
247439_at |
AT5G62670
|
AHA11, H(+)-ATPase 11, HA11, H(+)-ATPase 11 |
-0.263 |
-0.005 |
| 409 |
250245_at |
AT5G13690
|
CYL1, CYCLOPS 1, NAGLU, N-ACETYL-GLUCOSAMINIDASE |
-0.263 |
0.026 |
| 410 |
262286_at |
AT1G68585
|
unknown |
-0.262 |
-0.268 |
| 411 |
267549_at |
AT2G32640
|
[Lycopene beta/epsilon cyclase protein] |
-0.261 |
-0.051 |
| 412 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
-0.260 |
0.053 |
| 413 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
-0.259 |
-0.058 |
| 414 |
254328_at |
AT4G22570
|
APT3, adenine phosphoribosyl transferase 3 |
-0.259 |
-0.056 |
| 415 |
248558_at |
AT5G49990
|
[Xanthine/uracil permease family protein] |
-0.258 |
-0.168 |
| 416 |
262937_at |
AT1G79560
|
EMB1047, EMBRYO DEFECTIVE 1047, EMB156, EMBRYO DEFECTIVE 156, EMB36, EMBRYO DEFECTIVE 36, FTSH12, FTSH protease 12 |
-0.258 |
-0.039 |
| 417 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.257 |
-0.253 |
| 418 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.257 |
-0.109 |
| 419 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.256 |
-0.170 |
| 420 |
263596_at |
AT2G01900
|
[DNAse I-like superfamily protein] |
-0.255 |
-0.197 |
| 421 |
257197_at |
AT3G23800
|
SBP3, selenium-binding protein 3 |
-0.255 |
0.058 |
| 422 |
257533_at |
AT3G10840
|
[alpha/beta-Hydrolases superfamily protein] |
-0.253 |
-0.048 |
| 423 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.252 |
-0.129 |
| 424 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
-0.252 |
0.120 |
| 425 |
261230_at |
AT1G20010
|
TUB5, tubulin beta-5 chain |
-0.252 |
-0.028 |
| 426 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.252 |
0.061 |
| 427 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
-0.251 |
-0.079 |
| 428 |
251261_at |
AT3G62110
|
[Pectin lyase-like superfamily protein] |
-0.251 |
-0.021 |
| 429 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
-0.250 |
0.214 |