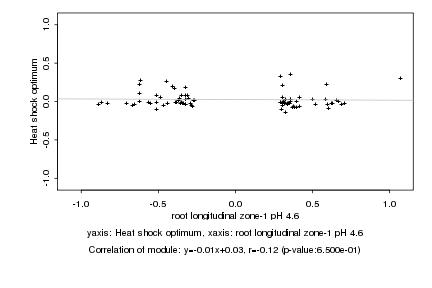
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
root longitudinal zone-1 pH 4.6 |
Log2 signal ratio
Heat shock optimum |
| 1 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
1.068 |
0.300 |
| 2 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
0.703 |
-0.020 |
| 3 |
249304_at |
AT5G41480
|
ATDFA, A. THALIANA DHFS-FPGS HOMOLOG A, DFA, DHFS-FPGS HOMOLOG A, EMB9, EMBRYO DEFECTIVE 9, GLA1, GLOBULAR ARREST1 |
0.682 |
-0.026 |
| 4 |
265234_at |
AT2G07721
|
unknown |
0.665 |
0.012 |
| 5 |
252689_at |
AT3G44030
|
[pseudogene] |
0.654 |
0.018 |
| 6 |
264301_at |
AT1G78780
|
[pathogenesis-related family protein] |
0.625 |
-0.025 |
| 7 |
245165_at |
AT2G33180
|
unknown |
0.618 |
-0.017 |
| 8 |
262745_at |
AT1G28600
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.600 |
-0.083 |
| 9 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
0.594 |
-0.026 |
| 10 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.587 |
0.227 |
| 11 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
0.581 |
0.028 |
| 12 |
264101_at |
AT1G79000
|
ATHAC1, ARABIDOPSIS HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY 1, ATHPCAT2, ARABIDOPSIS THALIANA P300/CBP ACETYLTRANSFERASE-RELATED PROTEIN 2, HAC1, histone acetyltransferase of the CBP family 1, PCAT2, P300/CBP ACETYLTRANSFERASE-RELATED PROTEIN 2 |
0.514 |
-0.031 |
| 13 |
245495_at |
AT4G16400
|
unknown |
0.494 |
0.030 |
| 14 |
256059_at |
AT1G06990
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.413 |
-0.054 |
| 15 |
261110_at |
AT1G75440
|
UBC16, ubiquitin-conjugating enzyme 16 |
0.410 |
0.057 |
| 16 |
252715_x_at |
AT3G43900
|
unknown |
0.396 |
-0.066 |
| 17 |
254288_at |
AT4G22970
|
AESP, homolog of separase, ESP, EXTRA SPINDLE POLES, ESP, homolog of separase, RSW4, RADIALLY SWOLLEN 4 |
0.393 |
0.011 |
| 18 |
256381_at |
AT1G66850
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.381 |
-0.070 |
| 19 |
266250_at |
AT2G27870
|
[similar to RNase H domain-containing protein [Arabidopsis thaliana] (TAIR:AT2G22350.1)] |
0.373 |
-0.055 |
| 20 |
265242_at |
AT2G07705
|
unknown |
0.367 |
-0.066 |
| 21 |
255923_at |
AT1G22180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.357 |
0.010 |
| 22 |
246015_at |
AT5G10700
|
[Peptidyl-tRNA hydrolase II (PTH2) family protein] |
0.356 |
0.031 |
| 23 |
266837_x_at |
AT2G25990
|
unknown |
0.356 |
0.033 |
| 24 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.355 |
0.360 |
| 25 |
250374_at |
AT5G11530
|
EMF1, embryonic flower 1 |
0.349 |
-0.024 |
| 26 |
265860_at |
AT2G01810
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.347 |
-0.018 |
| 27 |
256208_at |
AT1G50930
|
unknown |
0.341 |
-0.022 |
| 28 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
0.337 |
-0.021 |
| 29 |
265472_at |
AT2G15580
|
[RING/U-box superfamily protein] |
0.333 |
-0.031 |
| 30 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
0.324 |
0.030 |
| 31 |
253264_at |
AT4G33950
|
ATOST1, OPEN STOMATA 1, OST1, OPEN STOMATA 1, P44, SNRK2-6, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-6, SNRK2.6, SNF1-RELATED PROTEIN KINASE 2.6, SRK2E |
0.319 |
-0.137 |
| 32 |
266026_at |
AT2G05980
|
[non-LTR retrotransposon family (LINE), has a 1.0e-42 P-value blast match to GB:NP_038607 L1 repeat, Tf subfamily, member 9 (LINE-element) (Mus musculus)] |
0.316 |
-0.008 |
| 33 |
265176_at |
AT1G23520
|
unknown |
0.312 |
-0.015 |
| 34 |
252210_at |
AT3G50410
|
OBP1, OBF binding protein 1 |
0.308 |
0.012 |
| 35 |
257224_at |
AT3G27870
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.304 |
0.057 |
| 36 |
257331_at |
ATMG01330
|
ORF107H |
0.303 |
-0.016 |
| 37 |
251047_at |
AT5G02390
|
unknown |
0.302 |
-0.012 |
| 38 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.302 |
0.213 |
| 39 |
248763_at |
AT5G47550
|
[Cystatin/monellin superfamily protein] |
0.300 |
-0.040 |
| 40 |
267081_at |
AT2G41210
|
PIP5K5, phosphatidylinositol- 4-phosphate 5-kinase 5 |
0.295 |
-0.102 |
| 41 |
252522_at |
AT3G46390
|
unknown |
0.290 |
-0.011 |
| 42 |
244960_at |
ATCG01020
|
RPL32, ribosomal protein L32 |
0.289 |
-0.012 |
| 43 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.289 |
0.328 |
| 44 |
266276_at |
AT2G29330
|
TRI, tropinone reductase |
-0.890 |
-0.030 |
| 45 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.869 |
-0.001 |
| 46 |
264896_at |
AT1G23210
|
AtGH9B6, glycosyl hydrolase 9B6, GH9B6, glycosyl hydrolase 9B6 |
-0.830 |
-0.014 |
| 47 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.710 |
-0.024 |
| 48 |
263352_at |
AT2G22080
|
unknown |
-0.669 |
-0.041 |
| 49 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
-0.655 |
-0.032 |
| 50 |
259290_at |
AT3G11520
|
CYC2, CYCLIN 2, CYCB1;3, CYCLIN B1;3 |
-0.627 |
0.115 |
| 51 |
245674_at |
AT1G56680
|
[Chitinase family protein] |
-0.625 |
0.001 |
| 52 |
266655_at |
AT2G25880
|
AtAUR2, ataurora2, AUR2, ataurora2 |
-0.623 |
0.229 |
| 53 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
-0.618 |
0.286 |
| 54 |
255044_at |
AT4G09680
|
ATCTC1, CTC1, conserved telomere maintenance component 1 |
-0.565 |
-0.009 |
| 55 |
266711_at |
AT2G46740
|
AtGulLO5, GulLO5, L -gulono-1,4-lactone ( L -GulL) oxidase 5 |
-0.557 |
-0.025 |
| 56 |
251723_at |
AT3G56230
|
[BTB/POZ domain-containing protein] |
-0.517 |
-0.091 |
| 57 |
262516_at |
AT1G17190
|
ATGSTU26, glutathione S-transferase tau 26, GSTU26, glutathione S-transferase tau 26 |
-0.516 |
0.080 |
| 58 |
245245_at |
AT1G44318
|
hemb2 |
-0.515 |
-0.011 |
| 59 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.488 |
0.057 |
| 60 |
262331_at |
AT1G64050
|
unknown |
-0.471 |
-0.041 |
| 61 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
-0.451 |
0.263 |
| 62 |
246005_at |
AT5G08415
|
LIP1, lipoyl synthase 1 |
-0.444 |
-0.017 |
| 63 |
257005_at |
AT3G14190
|
CMR1, COPPER MODIFIED RESISTANCE 1 |
-0.414 |
0.198 |
| 64 |
264377_at |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
-0.397 |
0.176 |
| 65 |
266123_at |
AT2G45180
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.389 |
-0.012 |
| 66 |
256193_at |
AT1G30200
|
[F-box family protein] |
-0.388 |
-0.010 |
| 67 |
257274_at |
AT3G14510
|
[Polyprenyl synthetase family protein] |
-0.372 |
0.017 |
| 68 |
249818_at |
AT5G23860
|
TUB8, tubulin beta 8 |
-0.364 |
0.044 |
| 69 |
255572_at |
AT4G01050
|
TROL, thylakoid rhodanese-like |
-0.359 |
-0.022 |
| 70 |
265033_at |
AT1G61520
|
LHCA3, photosystem I light harvesting complex gene 3 |
-0.355 |
-0.003 |
| 71 |
250109_at |
AT5G15230
|
GASA4, GAST1 protein homolog 4 |
-0.353 |
0.079 |
| 72 |
250269_at |
AT5G12970
|
[Calcium-dependent lipid-binding (CaLB domain) plant phosphoribosyltransferase family protein] |
-0.346 |
-0.016 |
| 73 |
255428_at |
AT4G03390
|
SRF3, STRUBBELIG-receptor family 3 |
-0.342 |
-0.024 |
| 74 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.330 |
-0.028 |
| 75 |
246378_at |
AT1G57620
|
CYB, CYTOPLASMIC BODIES |
-0.330 |
0.034 |
| 76 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.326 |
-0.035 |
| 77 |
251778_at |
AT3G55660
|
ATROPGEF6, ROPGEF6, ROP (rho of plants) guanine nucleotide exchange factor 6 |
-0.326 |
0.080 |
| 78 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
-0.325 |
0.191 |
| 79 |
265349_at |
AT2G22610
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.312 |
0.087 |
| 80 |
262496_at |
AT1G21790
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
-0.305 |
0.046 |
| 81 |
256349_at |
AT1G54890
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.293 |
-0.019 |
| 82 |
254522_at |
AT4G19980
|
unknown |
-0.290 |
-0.049 |
| 83 |
265048_at |
AT1G52050
|
[Mannose-binding lectin superfamily protein] |
-0.280 |
-0.055 |
| 84 |
255478_at |
AT4G02440
|
EID1, EMPFINDLICHER IM DUNKELROTEN LICHT 1 |
-0.278 |
0.021 |
| 85 |
252048_at |
AT3G52500
|
[Eukaryotic aspartyl protease family protein] |
-0.272 |
0.021 |