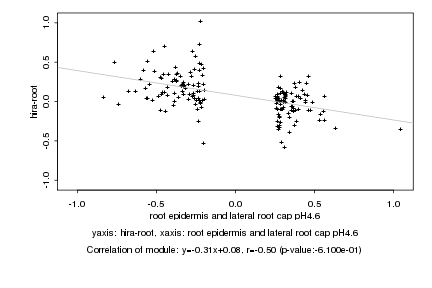
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
root epidermis and lateral root cap pH4.6 |
Log2 signal ratio
hira-root |
| 1 |
247902_at |
AT5G57350
|
AHA3, H(+)-ATPase 3, ATAHA3, ARABIDOPSIS THALIANA ARABIDOPSIS H(+)-ATPASE, HA3, H(+)-ATPase 3 |
1.042 |
-0.347 |
| 2 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
0.629 |
-0.341 |
| 3 |
259752_at |
AT1G71040
|
LPR2, Low Phosphate Root2 |
0.563 |
-0.232 |
| 4 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
0.560 |
0.066 |
| 5 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.551 |
-0.125 |
| 6 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
0.536 |
-0.156 |
| 7 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.526 |
-0.233 |
| 8 |
246850_at |
AT5G26860
|
LON1, lon protease 1, LON_ARA_ARA |
0.483 |
-0.009 |
| 9 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.472 |
-0.109 |
| 10 |
265560_at |
AT2G05520
|
ATGRP-3, GLYCINE-RICH PROTEIN 3, ATGRP3, ARABIDOPSIS GLYCINE RICH PROTEIN 3, GRP-3, glycine-rich protein 3, GRP3, GLYCINE-RICH PROTEIN 3 |
0.457 |
-0.112 |
| 11 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.457 |
0.320 |
| 12 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.450 |
0.081 |
| 13 |
246282_at |
AT4G36580
|
[AAA-type ATPase family protein] |
0.445 |
0.238 |
| 14 |
247799_at |
AT5G58840
|
[Subtilase family protein] |
0.441 |
0.016 |
| 15 |
263515_at |
AT2G21640
|
unknown |
0.439 |
-0.002 |
| 16 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
0.431 |
0.101 |
| 17 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.421 |
0.141 |
| 18 |
251029_at |
AT5G02050
|
[Mitochondrial glycoprotein family protein] |
0.408 |
0.051 |
| 19 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
0.400 |
0.243 |
| 20 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
0.394 |
0.067 |
| 21 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.393 |
-0.101 |
| 22 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.383 |
-0.249 |
| 23 |
245879_at |
AT5G09420
|
AtmtOM64, ATTOC64-V, translocon at the outer membrane of chloroplasts 64-V, MTOM64, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-V, OM64, outer membrane 64, TOC64-V, translocon at the outer membrane of chloroplasts 64-V |
0.382 |
0.075 |
| 24 |
247612_at |
AT5G60730
|
[Anion-transporting ATPase] |
0.378 |
0.186 |
| 25 |
260824_at |
AT1G06720
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.374 |
-0.113 |
| 26 |
254598_at |
AT4G18990
|
XTH29, xyloglucan endotransglucosylase/hydrolase 29 |
0.373 |
-0.295 |
| 27 |
264702_at |
AT1G70190
|
[Ribosomal protein L7/L12, oligomerisation] |
0.368 |
0.232 |
| 28 |
260585_at |
AT2G43650
|
EMB2777, EMBRYO DEFECTIVE 2777 |
0.366 |
-0.115 |
| 29 |
248049_at |
AT5G56090
|
COX15, cytochrome c oxidase 15 |
0.365 |
0.006 |
| 30 |
259465_at |
AT1G19030
|
unknown |
0.362 |
-0.082 |
| 31 |
264452_at |
AT1G10270
|
GRP23, glutamine-rich protein 23 |
0.358 |
-0.059 |
| 32 |
253777_at |
AT4G28450
|
[nucleotide binding] |
0.356 |
0.005 |
| 33 |
254079_at |
AT4G25730
|
[FtsJ-like methyltransferase family protein] |
0.351 |
-0.052 |
| 34 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.350 |
-0.104 |
| 35 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.349 |
0.111 |
| 36 |
254710_at |
AT4G18050
|
ABCB9, ATP-binding cassette B9, PGP9, P-glycoprotein 9 |
0.342 |
-0.389 |
| 37 |
255283_at |
AT4G04620
|
ATG8B, autophagy 8b |
0.340 |
-0.202 |
| 38 |
245209_at |
AT5G12340
|
unknown |
0.334 |
-0.149 |
| 39 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
0.321 |
0.115 |
| 40 |
263147_at |
AT1G53980
|
[Ubiquitin-like superfamily protein] |
0.315 |
0.083 |
| 41 |
253534_at |
AT4G31500
|
ATR4, ALTERED TRYPTOPHAN REGULATION 4, CYP83B1, cytochrome P450, family 83, subfamily B, polypeptide 1, RED1, RED ELONGATED 1, RNT1, RUNT 1, SUR2, SUPERROOT 2 |
0.314 |
-0.004 |
| 42 |
252355_at |
AT3G48250
|
BIR6, Buthionine sulfoximine-insensitive roots 6 |
0.314 |
0.092 |
| 43 |
250087_at |
AT5G17270
|
[Protein prenylyltransferase superfamily protein] |
0.313 |
0.040 |
| 44 |
245164_at |
AT2G33210
|
HSP60-2, heat shock protein 60-2 |
0.312 |
0.071 |
| 45 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.309 |
-0.572 |
| 46 |
249354_at |
AT5G40480
|
EMB3012, embryo defective 3012 |
0.308 |
0.052 |
| 47 |
262459_at |
AT1G50400
|
[Eukaryotic porin family protein] |
0.306 |
0.011 |
| 48 |
266782_at |
AT2G29120
|
ATGLR2.7, glutamate receptor 2.7, GLR2.7, GLUTAMATE RECEPTOR 2.7, GLR2.7, glutamate receptor 2.7 |
0.304 |
0.017 |
| 49 |
253175_at |
AT4G35050
|
MSI3, MULTICOPY SUPPRESSOR OF IRA1 3, NFC3, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 3 |
0.303 |
0.124 |
| 50 |
259444_at |
AT1G02370
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.303 |
0.111 |
| 51 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.299 |
-0.009 |
| 52 |
253942_at |
AT4G27010
|
EMB2788, EMBRYO DEFECTIVE 2788 |
0.298 |
-0.073 |
| 53 |
250679_at |
AT5G06550
|
JMJ22, Jumonji domain-containing protein 22 |
0.296 |
0.134 |
| 54 |
257058_at |
AT3G15352
|
ATCOX17, ARABIDOPSIS THALIANA CYTOCHROME C OXIDASE 17, COX17, cytochrome c oxidase 17 |
0.293 |
-0.101 |
| 55 |
258094_at |
AT3G14690
|
CYP72A15, cytochrome P450, family 72, subfamily A, polypeptide 15 |
0.291 |
-0.095 |
| 56 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.290 |
-0.511 |
| 57 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
0.287 |
0.019 |
| 58 |
266712_at |
AT2G46750
|
AtGulLO2, GulLO2, L -gulono-1,4-lactone ( L -GulL) oxidase 2 |
0.285 |
0.107 |
| 59 |
259422_at |
AT1G13810
|
[Restriction endonuclease, type II-like superfamily protein] |
0.283 |
0.108 |
| 60 |
247282_at |
AT5G64240
|
AtMC3, metacaspase 3, AtMCP1a, metacaspase 1a, MC3, metacaspase 3, MCP1a, metacaspase 1a |
0.283 |
0.173 |
| 61 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.282 |
-0.266 |
| 62 |
260641_at |
AT1G53200
|
unknown |
0.282 |
-0.089 |
| 63 |
263198_at |
AT1G53990
|
GLIP3, GDSL-motif lipase 3 |
0.281 |
0.322 |
| 64 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
0.280 |
-0.028 |
| 65 |
261168_at |
AT1G04945
|
[HIT-type Zinc finger family protein] |
0.278 |
0.006 |
| 66 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.275 |
-0.184 |
| 67 |
248495_at |
AT5G50780
|
[Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase family protein] |
0.274 |
-0.326 |
| 68 |
265151_at |
AT1G51340
|
[MATE efflux family protein] |
0.273 |
-0.293 |
| 69 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.273 |
-0.194 |
| 70 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.271 |
-0.351 |
| 71 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.269 |
0.186 |
| 72 |
247855_at |
AT5G58210
|
[hydroxyproline-rich glycoprotein family protein] |
0.268 |
0.016 |
| 73 |
249830_at |
AT5G23300
|
PYRD, pyrimidine d |
0.268 |
0.049 |
| 74 |
258179_at |
AT3G21690
|
[MATE efflux family protein] |
0.267 |
-0.160 |
| 75 |
255793_at |
AT2G33250
|
unknown |
0.266 |
0.052 |
| 76 |
267299_at |
AT2G30150
|
[UDP-Glycosyltransferase superfamily protein] |
0.264 |
0.098 |
| 77 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.264 |
0.036 |
| 78 |
264774_at |
AT1G22890
|
unknown |
0.263 |
0.067 |
| 79 |
248984_at |
AT5G45140
|
NRPC2, nuclear RNA polymerase C2 |
0.262 |
-0.090 |
| 80 |
245363_at |
AT4G15120
|
[VQ motif-containing protein] |
0.262 |
-0.253 |
| 81 |
252946_at |
AT4G39235
|
unknown |
0.261 |
0.016 |
| 82 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.261 |
-0.306 |
| 83 |
259008_at |
AT3G09390
|
ATMT-1, ARABIDOPSIS THALIANA METALLOTHIONEIN-1, ATMT-K, ARABIDOPSIS THALIANA METALLOTHIONEIN-K, MT2A, metallothionein 2A |
0.261 |
0.012 |
| 84 |
267004_at |
AT2G34260
|
WDR55, human WDR55 (WD40 repeat) homolog |
0.260 |
0.024 |
| 85 |
252246_at |
AT3G49725
|
[GTP-binding protein, HflX] |
0.259 |
-0.084 |
| 86 |
250787_at |
AT5G05560
|
APC1, anaphase promoting complex 1, EMB2771, EMBRYO DEFECTIVE 2771 |
0.257 |
-0.016 |
| 87 |
255317_at |
AT4G04180
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.254 |
-0.081 |
| 88 |
263843_at |
AT2G37020
|
[Translin family protein] |
0.253 |
0.050 |
| 89 |
257487_at |
AT1G71850
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
0.253 |
0.074 |
| 90 |
254949_at |
AT4G11020
|
unknown |
-0.837 |
0.055 |
| 91 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.766 |
0.498 |
| 92 |
257720_at |
AT3G18450
|
[PLAC8 family protein] |
-0.744 |
-0.029 |
| 93 |
256350_at |
AT1G54940
|
GUX4, Glucuronic Acid Substitution of Xylan 4, PGSIP4, plant glycogenin-like starch initiation protein 4 |
-0.682 |
0.131 |
| 94 |
264312_at |
AT1G70450
|
[Protein kinase superfamily protein] |
-0.635 |
0.129 |
| 95 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-0.601 |
0.289 |
| 96 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
-0.586 |
0.405 |
| 97 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
-0.573 |
0.166 |
| 98 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.565 |
0.051 |
| 99 |
264617_at |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
-0.562 |
0.048 |
| 100 |
249868_at |
AT5G23030
|
TET12, tetraspanin12 |
-0.558 |
0.512 |
| 101 |
265666_at |
AT2G27440
|
|
-0.544 |
0.223 |
| 102 |
251364_at |
AT3G61300
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.527 |
0.020 |
| 103 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
-0.523 |
0.641 |
| 104 |
262564_at |
AT1G34330
|
[pseudogene] |
-0.518 |
0.384 |
| 105 |
260723_at |
AT1G48070
|
[Thioredoxin superfamily protein] |
-0.488 |
0.067 |
| 106 |
261158_at |
AT1G34500
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
-0.478 |
-0.110 |
| 107 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.475 |
0.311 |
| 108 |
262904_at |
AT1G59725
|
[DNAJ heat shock family protein] |
-0.473 |
0.097 |
| 109 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.468 |
0.294 |
| 110 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
-0.465 |
0.123 |
| 111 |
264896_at |
AT1G23210
|
AtGH9B6, glycosyl hydrolase 9B6, GH9B6, glycosyl hydrolase 9B6 |
-0.460 |
0.347 |
| 112 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
-0.449 |
0.701 |
| 113 |
260053_at |
AT1G78120
|
TPR12, tetratricopeptide repeat 12 |
-0.449 |
0.122 |
| 114 |
259150_at |
AT3G10320
|
[Glycosyltransferase family 61 protein] |
-0.446 |
-0.115 |
| 115 |
251835_at |
AT3G55180
|
[alpha/beta-Hydrolases superfamily protein] |
-0.435 |
0.185 |
| 116 |
261708_at |
AT1G32740
|
[SBP (S-ribonuclease binding protein) family protein] |
-0.433 |
0.082 |
| 117 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.427 |
0.354 |
| 118 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.400 |
0.260 |
| 119 |
247610_at |
AT5G60630
|
unknown |
-0.397 |
-0.039 |
| 120 |
249263_at |
AT5G41730
|
[Protein kinase family protein] |
-0.391 |
0.006 |
| 121 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.388 |
0.292 |
| 122 |
259650_at |
AT1G55290
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
-0.387 |
0.113 |
| 123 |
256283_at |
AT3G12540
|
unknown |
-0.382 |
0.445 |
| 124 |
254324_at |
AT4G22640
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.378 |
0.264 |
| 125 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
-0.375 |
0.346 |
| 126 |
265134_at |
AT1G51260
|
LPAT3, lysophosphatidyl acyltransferase 3 |
-0.373 |
0.274 |
| 127 |
259964_at |
AT1G53680
|
ATGSTU28, glutathione S-transferase TAU 28, GSTU28, glutathione S-transferase TAU 28 |
-0.368 |
0.368 |
| 128 |
255756_at |
AT1G19940
|
AtGH9B5, glycosyl hydrolase 9B5, GH9B5, glycosyl hydrolase 9B5 |
-0.363 |
0.063 |
| 129 |
254097_at |
AT4G25160
|
[U-box domain-containing protein kinase family protein] |
-0.352 |
0.323 |
| 130 |
251405_at |
AT3G60330
|
AHA7, H(+)-ATPase 7, HA7, H(+)-ATPase 7 |
-0.346 |
0.205 |
| 131 |
267128_at |
AT2G23620
|
ATMES1, ARABIDOPSIS THALIANA METHYL ESTERASE 1, MES1, methyl esterase 1 |
-0.344 |
0.203 |
| 132 |
259850_at |
AT1G72240
|
unknown |
-0.341 |
0.137 |
| 133 |
262898_at |
AT1G59850
|
[ARM repeat superfamily protein] |
-0.336 |
0.224 |
| 134 |
250632_at |
AT5G07450
|
CYCP4;3, cyclin p4;3 |
-0.334 |
0.216 |
| 135 |
257418_at |
AT1G30850
|
RSH4, root hair specific 4 |
-0.333 |
0.093 |
| 136 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.331 |
0.199 |
| 137 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.330 |
0.249 |
| 138 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.327 |
0.213 |
| 139 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.322 |
0.177 |
| 140 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
-0.300 |
0.225 |
| 141 |
253413_at |
AT4G33020
|
ATZIP9, ZIP9 |
-0.300 |
0.035 |
| 142 |
246024_at |
AT5G21130
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.291 |
0.101 |
| 143 |
256368_at |
AT1G66800
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.290 |
0.380 |
| 144 |
262789_at |
AT1G10810
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.282 |
0.330 |
| 145 |
264775_at |
AT1G22880
|
ATCEL5, ARABIDOPSIS THALIANA CELLULASE 5, ATGH9B4, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B4, CEL5, cellulase 5 |
-0.278 |
0.641 |
| 146 |
247216_at |
AT5G64860
|
DPE1, disproportionating enzyme |
-0.277 |
0.072 |
| 147 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.271 |
0.103 |
| 148 |
258421_at |
AT3G16690
|
AtSWEET16, SWEET16 |
-0.270 |
0.205 |
| 149 |
262004_at |
AT1G64480
|
CBL8, calcineurin B-like protein 8 |
-0.266 |
0.080 |
| 150 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-0.263 |
0.412 |
| 151 |
250109_at |
AT5G15230
|
GASA4, GAST1 protein homolog 4 |
-0.260 |
0.044 |
| 152 |
245604_at |
AT4G14290
|
[alpha/beta-Hydrolases superfamily protein] |
-0.256 |
-0.030 |
| 153 |
266011_at |
AT2G37440
|
[DNAse I-like superfamily protein] |
-0.254 |
0.576 |
| 154 |
254754_at |
AT4G13210
|
[Pectin lyase-like superfamily protein] |
-0.247 |
0.020 |
| 155 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
-0.245 |
-0.089 |
| 156 |
266596_at |
AT2G46150
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.243 |
0.137 |
| 157 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.240 |
0.191 |
| 158 |
257450_at |
AT1G10530
|
unknown |
-0.237 |
0.230 |
| 159 |
253017_at |
AT4G37970
|
ATCAD6, CAD6, cinnamyl alcohol dehydrogenase 6 |
-0.236 |
-0.250 |
| 160 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.236 |
0.047 |
| 161 |
266191_at |
AT2G39040
|
[Peroxidase superfamily protein] |
-0.233 |
0.725 |
| 162 |
257690_at |
AT3G12830
|
SAUR72, SMALL AUXIN UPREGULATED 72 |
-0.232 |
0.128 |
| 163 |
249202_at |
AT5G42580
|
CYP705A12, cytochrome P450, family 705, subfamily A, polypeptide 12 |
-0.230 |
0.484 |
| 164 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.229 |
0.014 |
| 165 |
266548_at |
AT2G35210
|
AGD10, MEE28, MATERNAL EFFECT EMBRYO ARREST 28, RPA, root and pollen arfgap |
-0.228 |
0.396 |
| 166 |
254781_at |
AT4G12840
|
unknown |
-0.227 |
-0.012 |
| 167 |
247477_at |
AT5G62340
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.225 |
1.026 |
| 168 |
261096_at |
AT1G62940
|
ACOS5, acyl-CoA synthetase 5 |
-0.223 |
-0.017 |
| 169 |
253936_at |
AT4G26880
|
[Stigma-specific Stig1 family protein] |
-0.220 |
0.480 |
| 170 |
260907_at |
AT1G02575
|
unknown |
-0.216 |
-0.071 |
| 171 |
260495_at |
AT2G41810
|
unknown |
-0.214 |
0.340 |
| 172 |
261246_at |
AT1G20080
|
ATSYTB, NTMC2T1.2, NTMC2TYPE1.2, SYT2, synaptotagmin 2, SYTB |
-0.211 |
0.015 |
| 173 |
249814_at |
AT5G23840
|
[MD-2-related lipid recognition domain-containing protein] |
-0.208 |
0.425 |
| 174 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.205 |
-0.521 |
| 175 |
261074_at |
AT1G07290
|
GONST2, golgi nucleotide sugar transporter 2 |
-0.203 |
0.228 |
| 176 |
267612_at |
AT2G26690
|
AtNPF6.2, NPF6.2, NRT1/ PTR family 6.2 |
-0.201 |
0.145 |
| 177 |
260412_at |
AT1G69830
|
AMY3, alpha-amylase-like 3, ATAMY3, ALPHA-AMYLASE-LIKE 3 |
-0.200 |
0.029 |