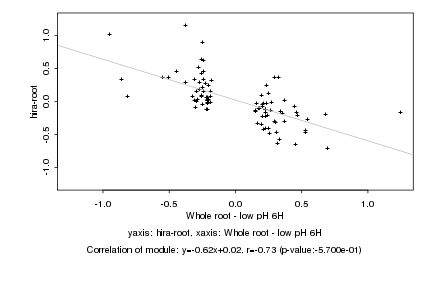
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Whole root - low pH 6H |
Log2 signal ratio
hira-root |
| 1 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
1.243 |
-0.162 |
| 2 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.691 |
-0.697 |
| 3 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.676 |
-0.195 |
| 4 |
257481_at |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
0.537 |
-0.259 |
| 5 |
260023_at |
AT1G30040
|
ATGA2OX2, gibberellin 2-oxidase, GA2OX2, gibberellin 2-oxidase, GA2OX2, GIBBERELLIN 2-OXIDASE 2 |
0.526 |
-0.461 |
| 6 |
263061_at |
AT2G18190
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.524 |
-0.431 |
| 7 |
262624_at |
AT1G06450
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.462 |
-0.197 |
| 8 |
247772_at |
AT5G58610
|
[PHD finger transcription factor, putative] |
0.455 |
-0.160 |
| 9 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.450 |
-0.647 |
| 10 |
251035_at |
AT5G02220
|
SMR4, SIAMESE-RELATED 4 |
0.445 |
-0.062 |
| 11 |
256852_at |
AT3G18610
|
ATNUC-L2, nucleolin like 2, NUC-L2, nucleolin like 2, PARLL1, PARALLEL1-LIKE 1 |
0.370 |
-0.301 |
| 12 |
250023_at |
AT5G18220
|
[O-Glycosyl hydrolases family 17 protein] |
0.369 |
0.021 |
| 13 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
0.354 |
-0.169 |
| 14 |
266597_at |
AT2G46130
|
ATWRKY43, WRKY DNA-BINDING PROTEIN 43, WRKY43, WRKY DNA-binding protein 43 |
0.335 |
-0.150 |
| 15 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.329 |
-0.572 |
| 16 |
260539_at |
AT2G43480
|
[Peroxidase superfamily protein] |
0.319 |
0.379 |
| 17 |
254200_at |
AT4G24110
|
unknown |
0.310 |
-0.627 |
| 18 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.304 |
-0.464 |
| 19 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.296 |
-0.315 |
| 20 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
0.294 |
-0.302 |
| 21 |
262353_at |
AT1G64210
|
[Leucine-rich repeat protein kinase family protein] |
0.291 |
0.373 |
| 22 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
0.265 |
-0.008 |
| 23 |
267276_at |
AT2G30130
|
ASL5, LBD12, PCK1, PEACOCK 1 |
0.259 |
-0.129 |
| 24 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.252 |
-0.475 |
| 25 |
262045_at |
AT1G80240
|
DGR1, DUF642 L-GalL responsive gene 1 |
0.246 |
0.125 |
| 26 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.243 |
-0.398 |
| 27 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.239 |
-0.199 |
| 28 |
253824_at |
AT4G27940
|
ATMTM1, ARABIDOPSIS MANGANESE TRACKING FACTOR FOR MITOCHONDRIAL SOD2, MTM1, manganese tracking factor for mitochondrial SOD2 |
0.234 |
-0.021 |
| 29 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
0.228 |
0.243 |
| 30 |
257140_at |
AT3G28910
|
ATMYB30, MYB30, myb domain protein 30 |
0.226 |
-0.406 |
| 31 |
253150_at |
AT4G35660
|
unknown |
0.225 |
-0.119 |
| 32 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.225 |
-0.159 |
| 33 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.225 |
-0.110 |
| 34 |
253203_at |
AT4G34710
|
ADC2, arginine decarboxylase 2, ATADC2, SPE2 |
0.221 |
-0.222 |
| 35 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.210 |
-0.412 |
| 36 |
267279_at |
AT2G19460
|
unknown |
0.208 |
-0.016 |
| 37 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.204 |
-0.222 |
| 38 |
255360_at |
AT4G03960
|
AtPFA-DSP4, PFA-DSP4, plant and fungi atypical dual-specificity phosphatase 4 |
0.202 |
-0.032 |
| 39 |
245909_at |
AT5G09380
|
[RNA polymerase III RPC4] |
0.198 |
-0.073 |
| 40 |
263937_at |
AT2G35910
|
[RING/U-box superfamily protein] |
0.194 |
0.093 |
| 41 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
0.193 |
-0.345 |
| 42 |
266963_at |
AT2G39450
|
ATMTP11, MTP11 |
0.180 |
-0.102 |
| 43 |
263436_at |
AT2G28690
|
unknown |
0.172 |
-0.101 |
| 44 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.161 |
-0.322 |
| 45 |
247670_at |
AT5G60190
|
[Cysteine proteinases superfamily protein] |
0.155 |
-0.021 |
| 46 |
259980_at |
AT1G76520
|
[Auxin efflux carrier family protein] |
0.150 |
-0.151 |
| 47 |
255719_at |
AT1G32080
|
AtLrgB, LrgB |
0.149 |
-0.130 |
| 48 |
247477_at |
AT5G62340
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.954 |
1.026 |
| 49 |
260495_at |
AT2G41810
|
unknown |
-0.862 |
0.340 |
| 50 |
267228_at |
AT2G43890
|
[Pectin lyase-like superfamily protein] |
-0.817 |
0.079 |
| 51 |
263610_at |
AT2G16230
|
[O-Glycosyl hydrolases family 17 protein] |
-0.557 |
0.365 |
| 52 |
246530_at |
AT5G15725
|
GLV9, GOLVEN 9 |
-0.508 |
0.379 |
| 53 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.448 |
0.456 |
| 54 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
-0.381 |
0.296 |
| 55 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.379 |
1.165 |
| 56 |
259222_at |
AT3G03680
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.331 |
0.079 |
| 57 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.315 |
0.335 |
| 58 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-0.314 |
0.028 |
| 59 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
-0.302 |
-0.089 |
| 60 |
258089_at |
AT3G14740
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.300 |
0.164 |
| 61 |
249788_at |
AT5G24330
|
ATXR6, ARABIDOPSIS TRITHORAX-RELATED PROTEIN 6, SDG34, SET DOMAIN PROTEIN 34 |
-0.295 |
0.005 |
| 62 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
-0.289 |
0.033 |
| 63 |
249836_at |
AT5G23420
|
HMGB6, high-mobility group box 6 |
-0.286 |
0.518 |
| 64 |
248511_at |
AT5G50375
|
CPI1, cyclopropyl isomerase |
-0.272 |
0.300 |
| 65 |
259042_at |
AT3G03450
|
RGL2, RGA-like 2 |
-0.272 |
0.188 |
| 66 |
265097_at |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
-0.262 |
0.098 |
| 67 |
264775_at |
AT1G22880
|
ATCEL5, ARABIDOPSIS THALIANA CELLULASE 5, ATGH9B4, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B4, CEL5, cellulase 5 |
-0.261 |
0.641 |
| 68 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
-0.261 |
0.080 |
| 69 |
248408_at |
AT5G51520
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.258 |
0.431 |
| 70 |
264682_at |
AT1G65570
|
[Pectin lyase-like superfamily protein] |
-0.251 |
0.908 |
| 71 |
252078_at |
AT3G51740
|
IMK2, inflorescence meristem receptor-like kinase 2 |
-0.249 |
-0.034 |
| 72 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.249 |
0.219 |
| 73 |
254284_at |
AT4G22910
|
CCS52A1, cell cycle switch protein 52 A1, FZR2, FIZZY-related 2 |
-0.248 |
0.340 |
| 74 |
250469_at |
AT5G10130
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.247 |
0.622 |
| 75 |
247741_at |
AT5G58960
|
GIL1, GRAVITROPIC IN THE LIGHT |
-0.244 |
0.155 |
| 76 |
267287_at |
AT2G23630
|
sks16, SKU5 similar 16 |
-0.243 |
0.469 |
| 77 |
265134_at |
AT1G51260
|
LPAT3, lysophosphatidyl acyltransferase 3 |
-0.233 |
0.274 |
| 78 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.226 |
-0.112 |
| 79 |
252266_at |
AT3G49630
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.224 |
0.063 |
| 80 |
251840_at |
AT3G54960
|
ATPDI1, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 1, ATPDIL1-3, PDI-like 1-3, PDI1, PROTEIN DISULFIDE ISOMERASE 1, PDIL1-3, PDI-like 1-3 |
-0.218 |
0.064 |
| 81 |
245404_at |
AT4G17610
|
[tRNA/rRNA methyltransferase (SpoU) family protein] |
-0.217 |
0.023 |
| 82 |
251067_at |
AT5G01910
|
unknown |
-0.215 |
-0.013 |
| 83 |
262758_at |
AT1G10780
|
[F-box/RNI-like superfamily protein] |
-0.213 |
0.028 |
| 84 |
255727_at |
AT1G25510
|
[Eukaryotic aspartyl protease family protein] |
-0.213 |
0.031 |
| 85 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.213 |
-0.018 |
| 86 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
-0.211 |
-0.118 |
| 87 |
258242_at |
AT3G27640
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.207 |
-0.006 |
| 88 |
263047_at |
AT2G17630
|
PSAT2, phosphoserine aminotransferase 2 |
-0.206 |
0.243 |
| 89 |
259151_at |
AT3G10310
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
-0.206 |
0.042 |
| 90 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.194 |
0.159 |
| 91 |
251046_at |
AT5G02370
|
[ATP binding microtubule motor family protein] |
-0.189 |
0.081 |
| 92 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
-0.189 |
-0.011 |
| 93 |
252361_at |
AT3G48490
|
unknown |
-0.188 |
0.325 |