|
probeID |
AGICode |
Annotation |
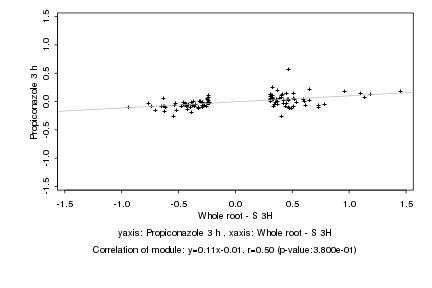
Log2 signal ratio
Whole root - S 3H |
Log2 signal ratio
Propiconazole 3 h |
| 1 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
1.448 |
0.188 |
| 2 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
1.186 |
0.126 |
| 3 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
1.130 |
0.077 |
| 4 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
1.096 |
0.151 |
| 5 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.959 |
0.180 |
| 6 |
264912_at |
AT1G60750
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.777 |
-0.040 |
| 7 |
253044_at |
AT4G37290
|
unknown |
0.730 |
-0.061 |
| 8 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.726 |
-0.101 |
| 9 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.646 |
0.031 |
| 10 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.645 |
0.227 |
| 11 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.610 |
-0.060 |
| 12 |
260540_at |
AT2G43500
|
NLP8, NIN-like protein 8 |
0.603 |
0.015 |
| 13 |
256762_at |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
0.590 |
0.053 |
| 14 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.536 |
-0.010 |
| 15 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.514 |
0.038 |
| 16 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
0.507 |
-0.076 |
| 17 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.503 |
0.154 |
| 18 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.503 |
0.060 |
| 19 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.490 |
-0.118 |
| 20 |
251824_at |
AT3G55090
|
ABCG16, ATP-binding cassette G16 |
0.472 |
-0.115 |
| 21 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
0.464 |
0.044 |
| 22 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
0.463 |
0.031 |
| 23 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
0.463 |
0.578 |
| 24 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
0.459 |
-0.098 |
| 25 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.456 |
0.052 |
| 26 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.447 |
0.142 |
| 27 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.447 |
-0.018 |
| 28 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
0.440 |
-0.084 |
| 29 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.420 |
0.022 |
| 30 |
254289_at |
AT4G22980
|
unknown |
0.416 |
-0.025 |
| 31 |
245560_at |
AT4G15480
|
UGT84A1 |
0.411 |
0.127 |
| 32 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.404 |
-0.249 |
| 33 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.404 |
0.075 |
| 34 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.398 |
0.114 |
| 35 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.379 |
0.056 |
| 36 |
248400_at |
AT5G52020
|
[Integrase-type DNA-binding superfamily protein] |
0.368 |
-0.040 |
| 37 |
252661_at |
AT3G44450
|
unknown |
0.368 |
0.196 |
| 38 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.365 |
0.004 |
| 39 |
264745_at |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
0.356 |
0.039 |
| 40 |
246584_at |
AT5G14730
|
unknown |
0.352 |
-0.011 |
| 41 |
267276_at |
AT2G30130
|
ASL5, LBD12, PCK1, PEACOCK 1 |
0.337 |
-0.045 |
| 42 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.331 |
0.045 |
| 43 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
0.330 |
-0.078 |
| 44 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
0.322 |
0.062 |
| 45 |
262653_at |
AT1G14130
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.322 |
0.038 |
| 46 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.322 |
0.113 |
| 47 |
250770_at |
AT5G05390
|
LAC12, laccase 12 |
0.320 |
0.265 |
| 48 |
245401_at |
AT4G17670
|
unknown |
0.319 |
0.072 |
| 49 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.312 |
0.115 |
| 50 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.308 |
0.009 |
| 51 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.307 |
0.050 |
| 52 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.307 |
0.137 |
| 53 |
245254_at |
AT4G14680
|
APS3 |
0.306 |
0.066 |
| 54 |
248381_at |
AT5G51830
|
[pfkB-like carbohydrate kinase family protein] |
0.305 |
0.103 |
| 55 |
265479_at |
AT2G15760
|
unknown |
0.304 |
0.037 |
| 56 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.942 |
-0.103 |
| 57 |
261567_at |
AT1G33055
|
unknown |
-0.765 |
-0.033 |
| 58 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-0.740 |
-0.073 |
| 59 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.703 |
-0.153 |
| 60 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
-0.657 |
-0.085 |
| 61 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-0.635 |
-0.084 |
| 62 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
-0.634 |
0.070 |
| 63 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
-0.630 |
-0.159 |
| 64 |
259316_at |
AT3G01175
|
unknown |
-0.628 |
-0.073 |
| 65 |
263970_at |
AT2G42850
|
CYP718, cytochrome P450, family 718 |
-0.622 |
-0.093 |
| 66 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.548 |
-0.254 |
| 67 |
248432_at |
AT5G51390
|
unknown |
-0.537 |
-0.064 |
| 68 |
259017_at |
AT3G07310
|
unknown |
-0.533 |
-0.033 |
| 69 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.524 |
-0.152 |
| 70 |
252036_at |
AT3G52070
|
unknown |
-0.477 |
-0.076 |
| 71 |
252882_at |
AT4G39675
|
unknown |
-0.463 |
-0.036 |
| 72 |
263265_at |
AT2G38820
|
unknown |
-0.460 |
-0.010 |
| 73 |
266140_at |
AT2G28120
|
[Major facilitator superfamily protein] |
-0.440 |
-0.026 |
| 74 |
250664_at |
AT5G07080
|
[HXXXD-type acyl-transferase family protein] |
-0.436 |
-0.083 |
| 75 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
-0.435 |
-0.076 |
| 76 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
-0.434 |
-0.061 |
| 77 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.434 |
-0.041 |
| 78 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.430 |
-0.137 |
| 79 |
254573_at |
AT4G19420
|
[Pectinacetylesterase family protein] |
-0.417 |
-0.038 |
| 80 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
-0.398 |
-0.105 |
| 81 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
-0.392 |
-0.183 |
| 82 |
249493_at |
AT5G39080
|
[HXXXD-type acyl-transferase family protein] |
-0.387 |
-0.001 |
| 83 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
-0.383 |
-0.068 |
| 84 |
247005_at |
AT5G67520
|
adenosine-5'-phosphosulfate (APS) kinase 4 |
-0.381 |
-0.054 |
| 85 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.369 |
0.012 |
| 86 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
-0.363 |
-0.082 |
| 87 |
248795_at |
AT5G47390
|
MYBH, MYB hypocotyl elongation-related |
-0.359 |
-0.043 |
| 88 |
252991_at |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
-0.332 |
-0.121 |
| 89 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.331 |
-0.097 |
| 90 |
250938_at |
AT5G03180
|
[RING/U-box superfamily protein] |
-0.317 |
0.004 |
| 91 |
260968_at |
AT1G12250
|
[Pentapeptide repeat-containing protein] |
-0.315 |
0.004 |
| 92 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.298 |
-0.061 |
| 93 |
264247_at |
AT1G60160
|
[Potassium transporter family protein] |
-0.298 |
-0.011 |
| 94 |
247072_at |
AT5G66490
|
unknown |
-0.296 |
-0.005 |
| 95 |
262355_at |
AT1G72820
|
Mitochondrial substrate carrier family protein |
-0.294 |
-0.089 |
| 96 |
266083_at |
AT2G37820
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.292 |
-0.032 |
| 97 |
247585_at |
AT5G60680
|
unknown |
-0.287 |
-0.076 |
| 98 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.277 |
-0.059 |
| 99 |
261667_at |
AT1G18460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.260 |
-0.076 |
| 100 |
256225_at |
AT1G56220
|
[Dormancy/auxin associated family protein] |
-0.254 |
-0.026 |
| 101 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
-0.254 |
0.040 |
| 102 |
261814_at |
AT1G08310
|
[alpha/beta-Hydrolases superfamily protein] |
-0.251 |
0.058 |
| 103 |
250107_at |
AT5G15330
|
ATSPX4, ARABIDOPSIS THALIANA SPX DOMAIN GENE 4, SPX4, SPX domain gene 4 |
-0.246 |
-0.007 |
| 104 |
264636_at |
AT1G65490
|
unknown |
-0.245 |
0.023 |
| 105 |
255068_at |
AT4G08920
|
ATCRY1, BLU1, BLUE LIGHT UNINHIBITED 1, CRY1, cryptochrome 1, HY4, ELONGATED HYPOCOTYL 4, OOP2, OUT OF PHASE 2 |
-0.245 |
-0.026 |
| 106 |
256622_at |
AT3G28920
|
AtHB34, homeobox protein 34, HB34, homeobox protein 34, ZHD9, ZINC FINGER HOMEODOMAIN 9 |
-0.240 |
0.118 |
| 107 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
-0.240 |
0.080 |
| 108 |
250017_at |
AT5G18140
|
DJC69, DNA J protein A69 |
-0.236 |
-0.034 |
| 109 |
253812_at |
AT4G28240
|
[Wound-responsive family protein] |
-0.236 |
-0.014 |