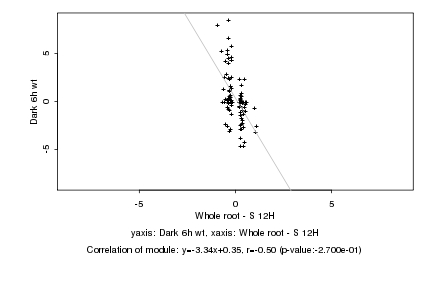
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Whole root - S 12H |
Log2 signal ratio
Dark 6h wt |
| 1 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
1.048 |
-2.583 |
| 2 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.999 |
-3.148 |
| 3 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.946 |
-0.658 |
| 4 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
0.575 |
-0.047 |
| 5 |
264912_at |
AT1G60750
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.509 |
-0.047 |
| 6 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.500 |
-0.256 |
| 7 |
265290_at |
AT2G22590
|
[UDP-Glycosyltransferase superfamily protein] |
0.477 |
-0.981 |
| 8 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.454 |
2.381 |
| 9 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.453 |
-4.206 |
| 10 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.441 |
-0.524 |
| 11 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.417 |
-2.701 |
| 12 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.392 |
-1.267 |
| 13 |
256762_at |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
0.385 |
-0.047 |
| 14 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.380 |
-4.683 |
| 15 |
245560_at |
AT4G15480
|
UGT84A1 |
0.376 |
-1.267 |
| 16 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
0.367 |
-0.047 |
| 17 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.361 |
-0.835 |
| 18 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
0.349 |
-0.104 |
| 19 |
253044_at |
AT4G37290
|
unknown |
0.325 |
-1.912 |
| 20 |
256454_at |
AT1G75280
|
[NmrA-like negative transcriptional regulator family protein] |
0.318 |
-2.265 |
| 21 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.312 |
0.890 |
| 22 |
260264_at |
AT1G68500
|
unknown |
0.296 |
1.749 |
| 23 |
254326_at |
AT4G22610
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.279 |
-0.047 |
| 24 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.279 |
0.533 |
| 25 |
249522_at |
AT5G38700
|
unknown |
0.278 |
-0.047 |
| 26 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.272 |
-2.343 |
| 27 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.272 |
0.001 |
| 28 |
254289_at |
AT4G22980
|
unknown |
0.271 |
-1.760 |
| 29 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.266 |
-2.897 |
| 30 |
267437_at |
AT2G19200
|
unknown |
0.263 |
-0.047 |
| 31 |
246584_at |
AT5G14730
|
unknown |
0.261 |
-0.452 |
| 32 |
262525_at |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
0.257 |
0.276 |
| 33 |
262552_at |
AT1G31350
|
KUF1, KAR-UP F-box 1 |
0.256 |
0.711 |
| 34 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
0.252 |
-1.128 |
| 35 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
0.251 |
0.114 |
| 36 |
264745_at |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
0.248 |
-4.658 |
| 37 |
260535_at |
AT2G43390
|
unknown |
0.247 |
-0.047 |
| 38 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.239 |
-0.047 |
| 39 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.234 |
-1.453 |
| 40 |
245401_at |
AT4G17670
|
unknown |
0.232 |
-2.433 |
| 41 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.231 |
-0.047 |
| 42 |
264710_at |
AT1G09790
|
COBL6, COBRA-like protein 6 precursor |
0.226 |
-0.047 |
| 43 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.224 |
-2.824 |
| 44 |
265422_at |
AT2G20800
|
NDB4, NAD(P)H dehydrogenase B4 |
0.223 |
-0.047 |
| 45 |
248621_at |
AT5G49350
|
[Glycine-rich protein family] |
0.221 |
-0.047 |
| 46 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
0.221 |
-3.853 |
| 47 |
262653_at |
AT1G14130
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.219 |
0.325 |
| 48 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.210 |
-0.532 |
| 49 |
251293_at |
AT3G61930
|
unknown |
0.208 |
-0.047 |
| 50 |
264906_at |
AT2G17270
|
MPT1, mitochondrial phosphate transporter 1, PHT3;3, phosphate transporter 3;3 |
0.202 |
-0.487 |
| 51 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.199 |
2.364 |
| 52 |
267070_at |
AT2G41000
|
[Chaperone DnaJ-domain superfamily protein] |
0.199 |
-0.519 |
| 53 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.198 |
-0.047 |
| 54 |
247634_at |
AT5G60520
|
[Late embryogenesis abundant (LEA) protein-related] |
0.196 |
-0.047 |
| 55 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.951 |
8.010 |
| 56 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
-0.726 |
5.325 |
| 57 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
-0.719 |
-0.047 |
| 58 |
259316_at |
AT3G01175
|
unknown |
-0.633 |
1.273 |
| 59 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.607 |
-0.047 |
| 60 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.596 |
2.514 |
| 61 |
267045_at |
AT2G34180
|
ATWL2, WPL4-LIKE 2, CIPK13, CBL-interacting protein kinase 13, SnRK3.7, SNF1-RELATED PROTEIN KINASE 3.7, WL2, WPL4-LIKE 2 |
-0.552 |
0.283 |
| 62 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
-0.544 |
4.197 |
| 63 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
-0.542 |
-2.329 |
| 64 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.472 |
2.869 |
| 65 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.464 |
5.355 |
| 66 |
252882_at |
AT4G39675
|
unknown |
-0.455 |
4.995 |
| 67 |
250664_at |
AT5G07080
|
[HXXXD-type acyl-transferase family protein] |
-0.437 |
0.166 |
| 68 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.436 |
4.931 |
| 69 |
260284_at |
AT1G80380
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.419 |
-0.583 |
| 70 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
-0.417 |
-2.529 |
| 71 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.394 |
2.434 |
| 72 |
258527_at |
AT3G06850
|
BCE2, DIN3, DARK INDUCIBLE 3, LTA1 |
-0.393 |
4.033 |
| 73 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-0.392 |
0.349 |
| 74 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.389 |
4.517 |
| 75 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
-0.381 |
2.433 |
| 76 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-0.379 |
8.565 |
| 77 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.378 |
-0.762 |
| 78 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.378 |
-0.047 |
| 79 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-0.374 |
6.641 |
| 80 |
252859_at |
AT4G39780
|
[Integrase-type DNA-binding superfamily protein] |
-0.363 |
-0.203 |
| 81 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.351 |
0.535 |
| 82 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
-0.344 |
-3.124 |
| 83 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
-0.332 |
-0.895 |
| 84 |
254648_at |
AT4G18550
|
AtDSEL, Arabidopsis thaliana DAD1-like seeding establishment-related lipase, DSEL, DAD1-like seeding establishment-related lipase |
-0.332 |
1.048 |
| 85 |
265387_at |
AT2G20670
|
unknown |
-0.331 |
2.357 |
| 86 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
-0.314 |
1.254 |
| 87 |
254573_at |
AT4G19420
|
[Pectinacetylesterase family protein] |
-0.312 |
1.187 |
| 88 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
-0.311 |
0.243 |
| 89 |
266276_at |
AT2G29330
|
TRI, tropinone reductase |
-0.304 |
-0.047 |
| 90 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.304 |
-2.841 |
| 91 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
-0.298 |
1.662 |
| 92 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
-0.286 |
1.663 |
| 93 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
-0.285 |
-0.031 |
| 94 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.270 |
-0.047 |
| 95 |
266670_at |
AT2G29740
|
UGT71C2, UDP-glucosyl transferase 71C2 |
-0.262 |
0.646 |
| 96 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
-0.260 |
0.464 |
| 97 |
258225_at |
AT3G15630
|
unknown |
-0.255 |
4.332 |
| 98 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
-0.251 |
5.817 |
| 99 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
-0.246 |
4.349 |
| 100 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
-0.236 |
4.679 |
| 101 |
245966_at |
AT5G19790
|
RAP2.11, related to AP2 11 |
-0.230 |
-0.047 |
| 102 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.225 |
1.397 |
| 103 |
266265_at |
AT2G29340
|
[NAD-dependent epimerase/dehydratase family protein] |
-0.223 |
-0.373 |
| 104 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
-0.223 |
-1.293 |
| 105 |
252834_at |
AT4G40070
|
[RING/U-box superfamily protein] |
-0.209 |
2.568 |
| 106 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-0.206 |
0.128 |
| 107 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
-0.200 |
-0.047 |