|
probeID |
AGICode |
Annotation |
Log2 signal ratio
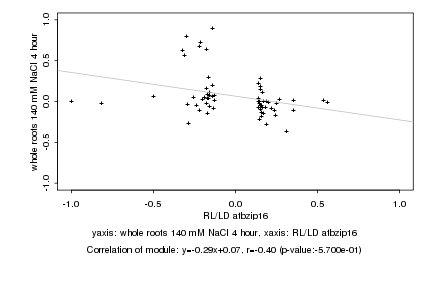
RL/LD atbzip16 |
Log2 signal ratio
whole roots 140 mM NaCl 4 hour |
| 1 |
255414_at |
AT4G03156
|
[small GTPase-related] |
0.557 |
-0.002 |
| 2 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.534 |
0.022 |
| 3 |
262010_at |
AT1G35612
|
[expressed protein] |
0.351 |
0.024 |
| 4 |
245859_at |
AT5G28290
|
ATNEK3, NIMA-related kinase 3, NEK3, NIMA-related kinase 3 |
0.348 |
-0.101 |
| 5 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
0.308 |
-0.367 |
| 6 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.263 |
0.034 |
| 7 |
253765_at |
AT4G28740
|
unknown |
0.247 |
-0.015 |
| 8 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
0.242 |
-0.169 |
| 9 |
259252_at |
AT3G07610
|
IBM1, increase in bonsai methylation 1 |
0.235 |
-0.104 |
| 10 |
266336_at |
AT2G32270
|
ZIP3, zinc transporter 3 precursor |
0.218 |
-0.074 |
| 11 |
259448_at |
AT1G13790
|
FDM4, factor of DNA methylation 4 |
0.196 |
-0.005 |
| 12 |
253187_at |
AT4G35280
|
DAZ2, DUO1-ACTIVATED ZINC FINGER 2 |
0.187 |
0.008 |
| 13 |
263610_at |
AT2G16230
|
[O-Glycosyl hydrolases family 17 protein] |
0.186 |
-0.274 |
| 14 |
262292_at |
AT1G27595
|
unknown |
0.180 |
-0.073 |
| 15 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.170 |
-0.146 |
| 16 |
248113_at |
AT5G55360
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
0.170 |
0.006 |
| 17 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
0.161 |
0.118 |
| 18 |
263748_at |
AT2G21480
|
[Malectin/receptor-like protein kinase family protein] |
0.160 |
-0.061 |
| 19 |
267588_at |
AT2G42060
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.156 |
-0.177 |
| 20 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.155 |
-0.131 |
| 21 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
0.155 |
-0.037 |
| 22 |
249497_at |
AT5G39220
|
[alpha/beta-Hydrolases superfamily protein] |
0.152 |
0.191 |
| 23 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
0.150 |
0.285 |
| 24 |
263673_at |
AT2G04800
|
unknown |
0.148 |
-0.038 |
| 25 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.148 |
0.159 |
| 26 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.148 |
-0.097 |
| 27 |
261596_at |
AT1G33080
|
[MATE efflux family protein] |
0.143 |
-0.210 |
| 28 |
252716_at |
AT3G43920
|
ATDCL3, DICER-LIKE 3, DCL3, dicer-like 3 |
0.143 |
-0.020 |
| 29 |
257550_at |
AT3G18460
|
[PLAC8 family protein] |
0.142 |
0.010 |
| 30 |
262046_at |
AT1G79960
|
ATOFP14, OFP14, ovate family protein 14 |
0.140 |
0.010 |
| 31 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.139 |
-0.065 |
| 32 |
247711_at |
AT5G59270
|
LecRK-II.2, L-type lectin receptor kinase II.2 |
0.139 |
0.039 |
| 33 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.138 |
0.222 |
| 34 |
260835_at |
AT1G06700
|
[Protein kinase superfamily protein] |
-1.004 |
0.007 |
| 35 |
254452_at |
AT4G21100
|
DDB1B, damaged DNA binding protein 1B |
-0.822 |
-0.022 |
| 36 |
262634_at |
AT1G06690
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.500 |
0.065 |
| 37 |
246241_at |
AT4G37050
|
AtPLAIVC, phospholipase A IVC, PLA V, PLAIII{beta}, patatin-related phospholipase III beta, PLP4, PATATIN-like protein 4 |
-0.323 |
0.636 |
| 38 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.314 |
0.574 |
| 39 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
-0.301 |
0.805 |
| 40 |
266634_at |
AT2G35530
|
AtbZIP16, bZIP16, basic region/leucine zipper transcription factor 16 |
-0.296 |
-0.032 |
| 41 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.290 |
-0.260 |
| 42 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
-0.257 |
0.058 |
| 43 |
262636_at |
AT1G06670
|
NIH, nuclear DEIH-boxhelicase |
-0.242 |
-0.043 |
| 44 |
246905_at |
AT5G25570
|
unknown |
-0.225 |
-0.099 |
| 45 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.220 |
0.685 |
| 46 |
257163_at |
AT3G24310
|
ATMYB71, MYB DOMAIN PROTEIN 71, MYB305, myb domain protein 305 |
-0.214 |
0.724 |
| 47 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.206 |
0.036 |
| 48 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.191 |
0.058 |
| 49 |
257718_at |
AT3G18400
|
anac058, NAC domain containing protein 58, NAC058, NAC domain containing protein 58 |
-0.180 |
-0.021 |
| 50 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.179 |
0.170 |
| 51 |
253494_at |
AT4G31830
|
unknown |
-0.177 |
0.637 |
| 52 |
253599_at |
AT4G30860
|
ASHR3, ASH1-related 3, SDG4, SET domain group 4 |
-0.176 |
0.098 |
| 53 |
249910_at |
AT5G22630
|
ADT5, arogenate dehydratase 5 |
-0.172 |
0.043 |
| 54 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.171 |
-0.146 |
| 55 |
266343_at |
AT2G01550
|
[non-LTR retrotransposon family (LINE), has a 4.9e-48 P-value blast match to GB:AAA39398 ORF2 (Mus musculus) (LINE-element)] |
-0.170 |
0.044 |
| 56 |
246908_at |
AT5G25610
|
ATRD22, RD22, RESPONSIVE TO DESSICATION 22 |
-0.165 |
0.295 |
| 57 |
249117_at |
AT5G43840
|
AT-HSFA6A, heat shock transcription factor A6A, HSFA6A, heat shock transcription factor A6A |
-0.164 |
0.075 |
| 58 |
251106_at |
AT5G01500
|
TAAC, thylakoid ATP/ADP carrier |
-0.160 |
-0.053 |
| 59 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
-0.159 |
0.120 |
| 60 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
-0.145 |
0.906 |
| 61 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
-0.144 |
0.065 |
| 62 |
254648_at |
AT4G18550
|
AtDSEL, Arabidopsis thaliana DAD1-like seeding establishment-related lipase, DSEL, DAD1-like seeding establishment-related lipase |
-0.140 |
0.201 |
| 63 |
265155_at |
AT1G30990
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.134 |
-0.085 |
| 64 |
252606_at |
AT3G45010
|
scpl48, serine carboxypeptidase-like 48 |
-0.132 |
0.075 |
| 65 |
256703_at |
AT3G30260
|
AGL79, AGAMOUS-like 79 |
-0.132 |
0.015 |