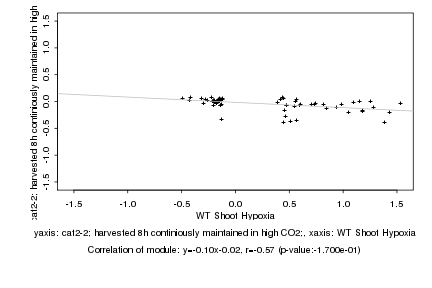
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
WT Shoot Hypoxia |
Log2 signal ratio
cat2-2; harvested 8h continiously maintained in high CO2; |
| 1 |
250464_at |
AT5G10040
|
unknown |
1.529 |
-0.026 |
| 2 |
247024_at |
AT5G66985
|
unknown |
1.424 |
-0.201 |
| 3 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
1.384 |
-0.391 |
| 4 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
1.276 |
-0.101 |
| 5 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
1.251 |
0.002 |
| 6 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
1.175 |
-0.186 |
| 7 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
1.172 |
-0.152 |
| 8 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
1.143 |
0.017 |
| 9 |
257925_at |
AT3G23170
|
unknown |
1.088 |
-0.008 |
| 10 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
1.041 |
-0.190 |
| 11 |
252882_at |
AT4G39675
|
unknown |
0.978 |
-0.050 |
| 12 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.938 |
-0.094 |
| 13 |
254200_at |
AT4G24110
|
unknown |
0.841 |
-0.123 |
| 14 |
250152_at |
AT5G15120
|
unknown |
0.813 |
-0.054 |
| 15 |
249384_at |
AT5G39890
|
unknown |
0.741 |
-0.029 |
| 16 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.731 |
-0.049 |
| 17 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.700 |
-0.055 |
| 18 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.599 |
-0.038 |
| 19 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.588 |
-0.057 |
| 20 |
260668_at |
AT1G19530
|
unknown |
0.566 |
-0.337 |
| 21 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.561 |
0.041 |
| 22 |
253404_at |
AT4G32840
|
PFK6, phosphofructokinase 6 |
0.553 |
0.008 |
| 23 |
261567_at |
AT1G33055
|
unknown |
0.545 |
-0.090 |
| 24 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.505 |
-0.356 |
| 25 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.471 |
-0.057 |
| 26 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.470 |
-0.063 |
| 27 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.463 |
-0.271 |
| 28 |
256366_at |
AT1G66880
|
[Protein kinase superfamily protein] |
0.452 |
-0.151 |
| 29 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.445 |
-0.389 |
| 30 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
0.443 |
0.068 |
| 31 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.428 |
0.075 |
| 32 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
0.417 |
0.045 |
| 33 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.384 |
-0.015 |
| 34 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
-0.494 |
0.062 |
| 35 |
264696_at |
AT1G70230
|
AXY4, ALTERED XYLOGLUCAN 4, TBL27, TRICHOME BIREFRINGENCE-LIKE 27 |
-0.433 |
0.028 |
| 36 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.420 |
0.075 |
| 37 |
252204_at |
AT3G50340
|
unknown |
-0.321 |
0.073 |
| 38 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.303 |
-0.032 |
| 39 |
253035_at |
AT4G38240
|
CGL, COMPLEX GLYCAN LESS, CGL1, COMPLEX GLYCAN LESS 1, GNTI, N-ACETYLGLUCOSAMINYLTRANSFERASE I |
-0.285 |
0.045 |
| 40 |
262745_at |
AT1G28600
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.260 |
0.021 |
| 41 |
248385_at |
AT5G51910
|
[TCP family transcription factor ] |
-0.230 |
0.076 |
| 42 |
263355_at |
AT2G22100
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.217 |
0.013 |
| 43 |
247147_at |
AT5G65630
|
GTE7, global transcription factor group E7 |
-0.214 |
0.018 |
| 44 |
247764_at |
AT5G59190
|
[subtilase family protein] |
-0.211 |
-0.067 |
| 45 |
254867_at |
AT4G12240
|
[zinc finger (C2H2 type) family protein] |
-0.205 |
0.032 |
| 46 |
257207_at |
AT3G14900
|
EMB3120, EMBRYO DEFECTIVE 3120 |
-0.198 |
0.026 |
| 47 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
-0.195 |
-0.004 |
| 48 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.178 |
0.016 |
| 49 |
245808_at |
AT1G58470
|
ATRBP1, RNA-binding protein 1, RBP1, RNA-binding protein 1 |
-0.177 |
-0.030 |
| 50 |
246566_at |
AT5G14940
|
[Major facilitator superfamily protein] |
-0.169 |
-0.015 |
| 51 |
247789_at |
AT5G58680
|
[ARM repeat superfamily protein] |
-0.163 |
0.050 |
| 52 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
-0.162 |
0.036 |
| 53 |
266801_at |
AT2G22870
|
EMB2001, embryo defective 2001 |
-0.157 |
0.044 |
| 54 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.155 |
0.067 |
| 55 |
251048_at |
AT5G02410
|
ALG10, homolog of yeast ALG10 |
-0.153 |
0.019 |
| 56 |
252164_at |
AT3G50620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.150 |
0.051 |
| 57 |
250424_at |
AT5G10550
|
GTE2, global transcription factor group E2 |
-0.145 |
0.022 |
| 58 |
265213_at |
AT1G05020
|
[ENTH/ANTH/VHS superfamily protein] |
-0.143 |
-0.059 |
| 59 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
-0.142 |
-0.064 |
| 60 |
252992_at |
AT4G38520
|
APD6, Arabidopsis Pp2c clade D 6 |
-0.140 |
0.032 |
| 61 |
262783_at |
AT1G10850
|
[Leucine-rich repeat protein kinase family protein] |
-0.139 |
0.046 |
| 62 |
264644_at |
AT1G08960
|
ATCAX11, CATION EXCHANGER 11, AtCXX5, Arabidopsis thaliana cation calcium exchanger 5, CAX11, cation exchanger 11, CCX5, cation calcium exchanger 5 |
-0.138 |
0.040 |
| 63 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
-0.135 |
-0.050 |
| 64 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.130 |
-0.331 |
| 65 |
258222_at |
AT3G15680
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-0.129 |
0.063 |
| 66 |
251075_at |
AT5G01890
|
PXC2, PXY/TDR-correlated 2 |
-0.125 |
0.042 |