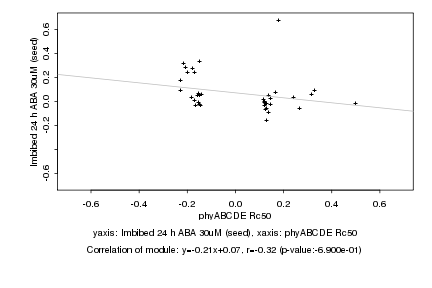
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
phyABCDE Rc50 |
Log2 signal ratio
Imbibed 24 h ABA 30uM (seed) |
| 1 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.495 |
-0.012 |
| 2 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
0.325 |
0.100 |
| 3 |
248218_at |
AT5G53710
|
unknown |
0.312 |
0.065 |
| 4 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.264 |
-0.056 |
| 5 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.238 |
0.041 |
| 6 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.177 |
0.684 |
| 7 |
264661_at |
AT1G09950
|
RAS1, RESPONSE TO ABA AND SALT 1 |
0.164 |
0.080 |
| 8 |
261642_at |
AT1G27680
|
APL2, ADPGLC-PPase large subunit |
0.144 |
-0.017 |
| 9 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.144 |
0.030 |
| 10 |
265898_at |
AT2G25690
|
unknown |
0.137 |
0.055 |
| 11 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
0.136 |
-0.084 |
| 12 |
252888_at |
AT4G39210
|
APL3 |
0.128 |
-0.150 |
| 13 |
265418_at |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
0.126 |
-0.011 |
| 14 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
0.125 |
-0.050 |
| 15 |
260945_at |
AT1G05950
|
unknown |
0.123 |
-0.016 |
| 16 |
254284_at |
AT4G22910
|
CCS52A1, cell cycle switch protein 52 A1, FZR2, FIZZY-related 2 |
0.121 |
-0.061 |
| 17 |
257954_at |
AT3G21760
|
HYR1, HYPOSTATIN RESISTANCE 1 |
0.119 |
-0.000 |
| 18 |
249087_at |
AT5G44210
|
ATERF-9, ERF DOMAIN PROTEIN- 9, ATERF9, ERF DOMAIN PROTEIN 9, ERF9, erf domain protein 9 |
0.117 |
-0.032 |
| 19 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
0.117 |
0.005 |
| 20 |
253318_at |
AT4G33970
|
[Leucine-rich repeat (LRR) family protein] |
0.116 |
0.021 |
| 21 |
247883_at |
AT5G57790
|
unknown |
-0.230 |
0.179 |
| 22 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
-0.229 |
0.096 |
| 23 |
257853_at |
AT3G12960
|
SMP1, seed maturation protein 1 |
-0.219 |
0.322 |
| 24 |
267036_at |
AT2G38465
|
unknown |
-0.209 |
0.291 |
| 25 |
249465_at |
AT5G39720
|
AIG2L, avirulence induced gene 2 like protein |
-0.200 |
0.250 |
| 26 |
259525_at |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
-0.185 |
0.038 |
| 27 |
259217_at |
AT3G03620
|
[MATE efflux family protein] |
-0.179 |
0.279 |
| 28 |
257457_at |
AT2G05430
|
[Ubiquitin-specific protease family C19-related protein] |
-0.172 |
0.009 |
| 29 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
-0.172 |
0.248 |
| 30 |
257334_at |
ATMG01370
|
ORF111D |
-0.167 |
-0.030 |
| 31 |
251291_at |
AT3G61900
|
SAUR33, SMALL AUXIN UPREGULATED RNA 33 |
-0.158 |
0.051 |
| 32 |
254200_at |
AT4G24110
|
unknown |
-0.157 |
-0.004 |
| 33 |
249772_at |
AT5G24130
|
unknown |
-0.154 |
0.074 |
| 34 |
250389_at |
AT5G11320
|
AtYUC4, YUC4, YUCCA4 |
-0.153 |
-0.024 |
| 35 |
245073_at |
AT2G23210
|
[UDP-Glycosyltransferase superfamily protein] |
-0.153 |
0.051 |
| 36 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
-0.152 |
-0.024 |
| 37 |
246299_at |
AT3G51810
|
AT3, ATEM1, ARABIDOPSIS THALIANA LATE EMBRYOGENESIS ABUNDANT 1, EM1, LATE EMBRYOGENESIS ABUNDANT 1, GEA1, GUANINE NUCLEOTIDE EXCHANGE FACTOR FOR ADP RIBOSYLATION FACTORS 1 |
-0.151 |
0.335 |
| 38 |
248038_at |
AT5G55980
|
[serine-rich protein-related] |
-0.147 |
-0.027 |
| 39 |
246188_at |
AT5G21050
|
[LOCATED IN: chloroplast] |
-0.145 |
0.061 |