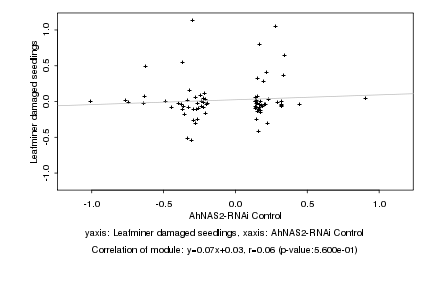
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
AhNAS2-RNAi Control |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
247491_at |
AT5G61880
|
[Protein Transporter, Pam16] |
0.905 |
0.043 |
| 2 |
253803_at |
AT4G28200
|
unknown |
0.441 |
-0.030 |
| 3 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.335 |
0.653 |
| 4 |
260429_at |
AT1G72450
|
JAZ6, jasmonate-zim-domain protein 6, TIFY11B, TIFY DOMAIN PROTEIN 11B |
0.329 |
0.375 |
| 5 |
254628_at |
AT4G18593
|
[dual specificity protein phosphatase-related] |
0.317 |
-0.053 |
| 6 |
257570_at |
AT3G13662
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.316 |
0.003 |
| 7 |
246264_at |
AT1G31814
|
FRL2, FRIGIDA like 2 |
0.315 |
-0.035 |
| 8 |
260709_at |
AT1G32500
|
ABCI7, ATP-binding cassette I7, ATNAP6, non-intrinsic ABC protein 6, NAP6, non-intrinsic ABC protein 6 |
0.314 |
-0.062 |
| 9 |
248579_at |
AT5G49880
|
AtMAD1, MAD1, mitotic arrest deficient 1, NES1, NITRIC OXIDE-INDUCED EARLY COTYLEDON SENESCENCE 1 |
0.287 |
-0.001 |
| 10 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.275 |
1.055 |
| 11 |
249980_at |
AT5G18870
|
NSH5, nucleoside hydrolase 5 |
0.228 |
0.032 |
| 12 |
265253_at |
AT2G02020
|
AtNPF8.4, AtPTR4, NPF8.4, NRT1/ PTR family 8.4, PTR4, peptide transporter 4 |
0.222 |
-0.304 |
| 13 |
259976_at |
AT1G76560
|
CP12-3, CP12 domain-containing protein 3 |
0.213 |
0.413 |
| 14 |
257224_at |
AT3G27870
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.206 |
-0.041 |
| 15 |
257509_at |
AT1G63190
|
[Cystatin/monellin superfamily protein] |
0.201 |
-0.041 |
| 16 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.190 |
0.288 |
| 17 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
0.185 |
-0.068 |
| 18 |
264843_at |
AT1G03400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.179 |
-0.053 |
| 19 |
245899_at |
AT5G11030
|
ALF4, ABERRANT LATERAL ROOT FORMATION 4 |
0.174 |
0.004 |
| 20 |
265472_at |
AT2G15580
|
[RING/U-box superfamily protein] |
0.171 |
-0.123 |
| 21 |
264121_at |
AT1G02280
|
ATTOC33, PPI1, PLASTID PROTEIN IMPORT 1, TOC33, translocon at the outer envelope membrane of chloroplasts 33 |
0.168 |
-0.144 |
| 22 |
246154_at |
AT5G19940
|
[Plastid-lipid associated protein PAP / fibrillin family protein] |
0.165 |
-0.041 |
| 23 |
264305_at |
AT1G78815
|
LSH7, LIGHT SENSITIVE HYPOCOTYLS 7 |
0.164 |
-0.074 |
| 24 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.162 |
0.806 |
| 25 |
261488_at |
AT1G14345
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.158 |
-0.416 |
| 26 |
266925_at |
AT2G45740
|
PEX11D, peroxin 11D |
0.157 |
-0.109 |
| 27 |
261710_at |
AT1G32730
|
unknown |
0.153 |
-0.138 |
| 28 |
245444_at |
AT4G16740
|
ATTPS03, terpene synthase 03, TPS03, terpene synthase 03 |
0.148 |
0.325 |
| 29 |
248886_at |
AT5G46110
|
APE2, ACCLIMATION OF PHOTOSYNTHESIS TO ENVIRONMENT 2, TPT, triose-phosphate ⁄ phosphate translocator |
0.147 |
-0.022 |
| 30 |
248379_at |
AT5G51700
|
ATRAR1, PBS2, PPHB SUSCEPTIBLE 2, RAR1, Required for Mla12 resistance 1, RPR2 |
0.147 |
0.084 |
| 31 |
245267_at |
AT4G14060
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.144 |
-0.022 |
| 32 |
256323_at |
AT1G54920
|
unknown |
0.143 |
-0.081 |
| 33 |
257823_at |
AT3G25190
|
[Vacuolar iron transporter (VIT) family protein] |
0.142 |
-0.074 |
| 34 |
257407_at |
AT1G27100
|
[Actin cross-linking protein] |
0.141 |
0.025 |
| 35 |
245792_at |
AT1G32100
|
ATPRR1, PRR1, pinoresinol reductase 1 |
0.141 |
-0.242 |
| 36 |
250920_at |
AT5G03390
|
unknown |
0.139 |
0.057 |
| 37 |
266600_at |
AT2G46070
|
ATMPK12, MAPK12, MPK12, mitogen-activated protein kinase 12 |
0.138 |
-0.056 |
| 38 |
252819_at |
AT3G42600
|
unknown |
0.135 |
0.007 |
| 39 |
261409_at |
AT1G07640
|
OBP2, URP3, UAS-TAGGED ROOT PATTERNING3 |
0.134 |
-0.088 |
| 40 |
247959_at |
AT5G57080
|
unknown |
0.134 |
0.014 |
| 41 |
263296_at |
AT2G38800
|
[Plant calmodulin-binding protein-related] |
-1.014 |
0.005 |
| 42 |
262442_at |
AT1G47420
|
SDH5, succinate dehydrogenase 5 |
-0.767 |
0.015 |
| 43 |
254355_at |
AT4G22380
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
-0.747 |
-0.005 |
| 44 |
250924_at |
AT5G03440
|
unknown |
-0.643 |
-0.016 |
| 45 |
251318_at |
AT3G61570
|
GC3, GOLGIN CANDIDATE 3, GDAP1, GRIP-related ARF-binding domain-containing protein 1 |
-0.638 |
0.072 |
| 46 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.627 |
0.498 |
| 47 |
252491_at |
AT3G46730
|
[NB-ARC domain-containing disease resistance protein] |
-0.490 |
0.011 |
| 48 |
247016_at |
AT5G66970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.449 |
-0.072 |
| 49 |
262841_at |
AT1G14810
|
[semialdehyde dehydrogenase family protein] |
-0.402 |
-0.021 |
| 50 |
264495_at |
AT1G27380
|
RIC2, ROP-interactive CRIB motif-containing protein 2 |
-0.378 |
-0.038 |
| 51 |
263754_at |
AT2G21510
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.374 |
0.556 |
| 52 |
256750_at |
AT3G27150
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.373 |
-0.106 |
| 53 |
250954_at |
AT5G03520
|
ATRAB-E1D, ARABIDOPSIS RAB HOMOLOG E1D, ATRAB8C, RAB GTPase homolog 8C, ATRABE1D, RAB-E1D, RAB HOMOLOG E1D, RAB8C, RAB GTPase homolog 8C |
-0.362 |
-0.056 |
| 54 |
253286_at |
AT4G34260
|
AXY8, ALTERED XYLOGLUCAN 8, FUC95A |
-0.357 |
-0.179 |
| 55 |
265464_at |
AT2G37080
|
RIP2, ROP interactive partner 2 |
-0.339 |
-0.511 |
| 56 |
256029_at |
AT1G34130
|
STT3B, staurosporin and temperature sensitive 3-like b |
-0.335 |
0.023 |
| 57 |
245891_at |
AT5G09220
|
AAP2, amino acid permease 2 |
-0.332 |
-0.083 |
| 58 |
266000_at |
AT2G24180
|
CYP71B6, cytochrome p450 71b6 |
-0.321 |
0.160 |
| 59 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
-0.306 |
-0.543 |
| 60 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.305 |
1.146 |
| 61 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.296 |
-0.254 |
| 62 |
250444_at |
AT5G10560
|
[Glycosyl hydrolase family protein] |
-0.295 |
-0.103 |
| 63 |
246797_at |
AT5G26790
|
unknown |
-0.281 |
-0.295 |
| 64 |
264657_at |
AT1G09100
|
RPT5B, 26S proteasome AAA-ATPase subunit RPT5B |
-0.278 |
0.059 |
| 65 |
256832_at |
AT3G22880
|
ARLIM15, ARABIDOPSIS HOMOLOG OF LILY MESSAGES INDUCED AT MEIOSIS 15, ATDMC1, ARABIDOPSIS THALIANA DISRUPTION OF MEIOTIC CONTROL 1, DMC1, DISRUPTION OF MEIOTIC CONTROL 1 |
-0.274 |
-0.107 |
| 66 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.271 |
-0.243 |
| 67 |
249040_at |
AT5G44320
|
[Eukaryotic translation initiation factor 3 subunit 7 (eIF-3)] |
-0.269 |
-0.022 |
| 68 |
267208_at |
AT2G30980
|
ASKdZeta, SHAGGY-related protein kinase dZeta, ATSK2-2, A. THALIANA SHAGGY-LIKE KINASE GROUP 2 2, ATSK23, SHAGGY-LIKE PROTEIN KINASE 23, BIL1, BIN2-LIKE 1, SKdZeta, SHAGGY-related protein kinase dZeta |
-0.259 |
-0.096 |
| 69 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
-0.250 |
0.089 |
| 70 |
258966_at |
AT3G10690
|
GYRA, DNA GYRASE A |
-0.240 |
-0.064 |
| 71 |
250233_at |
AT5G13460
|
IQD11, IQ-domain 11 |
-0.239 |
0.010 |
| 72 |
256145_at |
AT1G48750
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.225 |
-0.077 |
| 73 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
-0.225 |
-0.012 |
| 74 |
248053_at |
AT5G55810
|
AtNMNAT, nicotinate/nicotinamide mononucleotide adenyltransferase, NMNAT, nicotinate/nicotinamide mononucleotide adenyltransferase |
-0.225 |
0.045 |
| 75 |
257229_at |
AT3G16490
|
IQD26, IQ-domain 26 |
-0.222 |
0.125 |
| 76 |
247098_at |
AT5G66470
|
[RNA binding] |
-0.214 |
-0.154 |
| 77 |
251262_at |
AT3G62080
|
[SNF7 family protein] |
-0.210 |
0.033 |
| 78 |
257732_at |
AT3G18480
|
AtCASP, CCAAT-displacement protein alternatively spliced product, CASP, CCAAT-displacement protein alternatively spliced product |
-0.204 |
-0.032 |
| 79 |
266579_at |
AT2G23930
|
SNRNP-G, probable small nuclear ribonucleoprotein G |
-0.201 |
-0.018 |