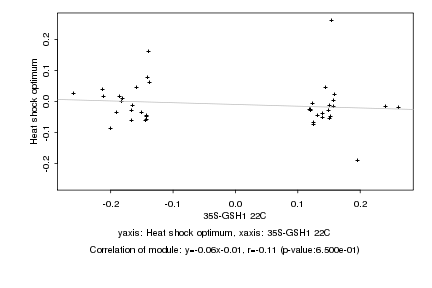
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35S-GSH1 22C |
Log2 signal ratio
Heat shock optimum |
| 1 |
256087_at |
AT1G20800
|
[F-box family protein] |
0.261 |
-0.017 |
| 2 |
248886_at |
AT5G46110
|
APE2, ACCLIMATION OF PHOTOSYNTHESIS TO ENVIRONMENT 2, TPT, triose-phosphate ⁄ phosphate translocator |
0.240 |
-0.014 |
| 3 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
0.195 |
-0.187 |
| 4 |
263418_at |
AT2G17210
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.158 |
0.023 |
| 5 |
266990_at |
AT2G39190
|
ATATH8 |
0.157 |
-0.016 |
| 6 |
252767_at |
AT3G42830
|
[RING/U-box superfamily protein] |
0.157 |
0.004 |
| 7 |
266613_at |
AT2G14900
|
[Gibberellin-regulated family protein] |
0.154 |
0.264 |
| 8 |
260403_at |
AT1G69810
|
ATWRKY36, WRKY36, WRKY DNA-binding protein 36 |
0.151 |
-0.048 |
| 9 |
265125_at |
AT1G55410
|
[pseudogene] |
0.150 |
-0.010 |
| 10 |
260646_at |
AT1G53340
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.150 |
-0.054 |
| 11 |
265115_at |
AT1G62450
|
[Immunoglobulin E-set superfamily protein] |
0.148 |
-0.027 |
| 12 |
258326_at |
AT3G22760
|
SOL1 |
0.143 |
0.048 |
| 13 |
265189_at |
AT1G23840
|
unknown |
0.139 |
-0.051 |
| 14 |
247719_at |
AT5G59305
|
unknown |
0.138 |
-0.036 |
| 15 |
259289_at |
AT3G11350
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.131 |
-0.043 |
| 16 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
0.125 |
-0.072 |
| 17 |
250658_at |
AT5G07040
|
[RING/U-box superfamily protein] |
0.125 |
-0.067 |
| 18 |
266973_at |
AT2G39620
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.123 |
-0.006 |
| 19 |
247844_at |
AT5G58080
|
ARR18, response regulator 18, RR18, response regulator 18 |
0.120 |
-0.026 |
| 20 |
251514_at |
AT3G59260
|
[pirin, putative] |
0.119 |
-0.025 |
| 21 |
266711_at |
AT2G46740
|
AtGulLO5, GulLO5, L -gulono-1,4-lactone ( L -GulL) oxidase 5 |
0.118 |
-0.025 |
| 22 |
263409_at |
AT2G04063
|
[glycine-rich protein] |
-0.261 |
0.029 |
| 23 |
258945_at |
AT3G10660
|
ATCPK2, CPK2, calmodulin-domain protein kinase cdpk isoform 2 |
-0.214 |
0.041 |
| 24 |
262470_at |
AT1G50180
|
[NB-ARC domain-containing disease resistance protein] |
-0.213 |
0.017 |
| 25 |
254122_at |
AT4G24510
|
CER2, ECERIFERUM 2, VC-2, VC2 |
-0.201 |
-0.085 |
| 26 |
250796_at |
AT5G05300
|
unknown |
-0.191 |
-0.035 |
| 27 |
254248_at |
AT4G23270
|
CRK19, cysteine-rich RLK (RECEPTOR-like protein kinase) 19 |
-0.186 |
0.018 |
| 28 |
245907_at |
AT5G09340
|
[Ubiquitin family protein] |
-0.184 |
0.003 |
| 29 |
252309_at |
AT3G49340
|
[Cysteine proteinases superfamily protein] |
-0.182 |
0.010 |
| 30 |
262625_at |
AT1G06440
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
-0.168 |
-0.061 |
| 31 |
257421_at |
AT1G12030
|
unknown |
-0.168 |
-0.028 |
| 32 |
251213_at |
AT3G62490
|
ASY2 |
-0.166 |
-0.012 |
| 33 |
254971_at |
AT4G10380
|
AtNIP5;1, NIP5;1, NOD26-like intrinsic protein 5;1, NLM6, NOD26-LIKE MIP 6, NLM8, NOD26-LIKE MIP 8 |
-0.159 |
0.048 |
| 34 |
254022_at |
AT4G25750
|
ABCG4, ATP-binding cassette G4 |
-0.152 |
-0.035 |
| 35 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
-0.145 |
-0.058 |
| 36 |
255203_at |
AT4G07510
|
unknown |
-0.144 |
-0.046 |
| 37 |
256273_at |
AT3G12090
|
TET6, tetraspanin6 |
-0.144 |
-0.056 |
| 38 |
249379_at |
AT5G40460
|
unknown |
-0.143 |
-0.043 |
| 39 |
245906_at |
AT5G11070
|
unknown |
-0.141 |
0.079 |
| 40 |
260028_at |
AT1G29980
|
unknown |
-0.140 |
0.163 |
| 41 |
251355_at |
AT3G61100
|
[Putative endonuclease or glycosyl hydrolase] |
-0.138 |
0.063 |