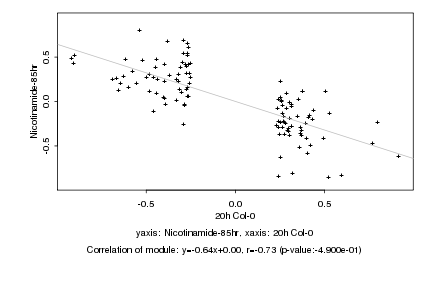
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
20h Col-0 |
Log2 signal ratio
Nicotinamide-85hr |
| 1 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.910 |
-0.608 |
| 2 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.795 |
-0.227 |
| 3 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.763 |
-0.472 |
| 4 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.592 |
-0.822 |
| 5 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
0.524 |
-0.132 |
| 6 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
0.521 |
-0.853 |
| 7 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
0.502 |
0.113 |
| 8 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.489 |
-0.407 |
| 9 |
257832_at |
AT3G26740
|
CCL, CCR-like |
0.432 |
-0.091 |
| 10 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.429 |
-0.201 |
| 11 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.418 |
-0.491 |
| 12 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.410 |
-0.156 |
| 13 |
246929_at |
AT5G25210
|
unknown |
0.404 |
-0.173 |
| 14 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
0.402 |
-0.575 |
| 15 |
260688_at |
AT1G17665
|
unknown |
0.393 |
-0.407 |
| 16 |
253322_at |
AT4G33980
|
unknown |
0.390 |
-0.238 |
| 17 |
262533_at |
AT1G17090
|
unknown |
0.374 |
0.118 |
| 18 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.374 |
0.124 |
| 19 |
253581_at |
AT4G30660
|
[Low temperature and salt responsive protein family] |
0.369 |
-0.323 |
| 20 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
0.369 |
-0.372 |
| 21 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
0.364 |
-0.282 |
| 22 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.361 |
-0.356 |
| 23 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
0.355 |
-0.513 |
| 24 |
267036_at |
AT2G38465
|
unknown |
0.353 |
0.034 |
| 25 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.345 |
-0.159 |
| 26 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.315 |
-0.802 |
| 27 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
0.314 |
-0.270 |
| 28 |
262296_at |
AT1G27630
|
CYCT1;3, cyclin T 1;3 |
0.314 |
-0.054 |
| 29 |
266839_at |
AT2G25930
|
ELF3, EARLY FLOWERING 3, PYK20 |
0.310 |
-0.032 |
| 30 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.309 |
-0.277 |
| 31 |
245600_at |
AT4G14230
|
unknown |
0.301 |
-0.189 |
| 32 |
249456_at |
AT5G39410
|
[Saccharopine dehydrogenase ] |
0.301 |
-0.009 |
| 33 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
0.299 |
-0.379 |
| 34 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
0.297 |
-0.335 |
| 35 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.295 |
-0.298 |
| 36 |
255953_at |
AT1G22070
|
TGA3, TGA1A-related gene 3 |
0.287 |
-0.317 |
| 37 |
250194_at |
AT5G14550
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.283 |
0.097 |
| 38 |
250987_at |
AT5G02860
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.281 |
-0.070 |
| 39 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.275 |
-0.244 |
| 40 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.273 |
-0.371 |
| 41 |
247043_at |
AT5G66880
|
SNRK2-3, SUCROSE NONFERMENTING 1 (SNF1)-RELATED PROTEIN KINASE 2-3, SNRK2.3, sucrose nonfermenting 1(SNF1)-related protein kinase 2.3, SRK2I |
0.273 |
-0.234 |
| 42 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
0.266 |
-0.161 |
| 43 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
0.264 |
-0.215 |
| 44 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
0.263 |
-0.129 |
| 45 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.259 |
-0.292 |
| 46 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.259 |
-0.040 |
| 47 |
251789_at |
AT3G55450
|
PBL1, PBS1-like 1 |
0.256 |
0.015 |
| 48 |
267280_at |
AT2G19450
|
ABX45, AS11, ATDGAT, Arabidopsis thaliana acyl-CoA:diacylglycerol acyltransferase, AtDGAT1, DGAT1, acyl-CoA:diacylglycerol acyltransferase 1, RDS1, TAG1, TRIACYLGLYCEROL BIOSYNTHESIS DEFECT 1 |
0.255 |
0.003 |
| 49 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
0.250 |
-0.626 |
| 50 |
249128_at |
AT5G43440
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.250 |
-0.234 |
| 51 |
245789_at |
AT1G32090
|
[early-responsive to dehydration stress protein (ERD4)] |
0.250 |
0.056 |
| 52 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.248 |
0.234 |
| 53 |
252468_at |
AT3G46970
|
ATPHS2, Arabidopsis thaliana alpha-glucan phosphorylase 2, PHS2, alpha-glucan phosphorylase 2 |
0.244 |
-0.368 |
| 54 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
0.241 |
-0.836 |
| 55 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
0.241 |
-0.222 |
| 56 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
0.237 |
-0.292 |
| 57 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
0.236 |
0.033 |
| 58 |
252464_at |
AT3G47160
|
[RING/U-box superfamily protein] |
0.233 |
-0.068 |
| 59 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
0.229 |
-0.263 |
| 60 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.923 |
0.490 |
| 61 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
-0.913 |
0.438 |
| 62 |
261263_at |
AT1G26790
|
[Dof-type zinc finger DNA-binding family protein] |
-0.907 |
0.526 |
| 63 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.694 |
0.249 |
| 64 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.671 |
0.267 |
| 65 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
-0.660 |
0.133 |
| 66 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.648 |
0.213 |
| 67 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
-0.632 |
0.288 |
| 68 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
-0.618 |
0.483 |
| 69 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
-0.603 |
0.162 |
| 70 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.579 |
0.338 |
| 71 |
247921_at |
AT5G57660
|
ATCOL5, BBX6, B-box domain protein 6, COL5, CONSTANS-like 5 |
-0.557 |
0.204 |
| 72 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.540 |
0.801 |
| 73 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
-0.522 |
0.466 |
| 74 |
265387_at |
AT2G20670
|
unknown |
-0.503 |
0.271 |
| 75 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.486 |
0.309 |
| 76 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
-0.483 |
0.115 |
| 77 |
250327_at |
AT5G12050
|
unknown |
-0.463 |
-0.108 |
| 78 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
-0.460 |
0.272 |
| 79 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.452 |
0.391 |
| 80 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
-0.445 |
0.484 |
| 81 |
264238_at |
AT1G54740
|
unknown |
-0.444 |
0.101 |
| 82 |
260956_at |
AT1G06040
|
BBX24, B-box domain protein 24, STO, SALT TOLERANCE |
-0.439 |
0.253 |
| 83 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
-0.405 |
0.054 |
| 84 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
-0.402 |
0.041 |
| 85 |
266363_at |
AT2G41250
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.401 |
0.422 |
| 86 |
251793_at |
AT3G55580
|
[Regulator of chromosome condensation (RCC1) family protein] |
-0.401 |
0.234 |
| 87 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.396 |
-0.032 |
| 88 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.384 |
0.684 |
| 89 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.370 |
0.298 |
| 90 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
-0.334 |
0.249 |
| 91 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
-0.333 |
0.019 |
| 92 |
252917_at |
AT4G38960
|
BBX19, B-box domain protein 19 |
-0.321 |
0.312 |
| 93 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.320 |
0.235 |
| 94 |
247474_at |
AT5G62280
|
unknown |
-0.319 |
0.145 |
| 95 |
253125_at |
AT4G36040
|
DJC23, DNA J protein C23, J11, DnaJ11 |
-0.310 |
0.390 |
| 96 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
-0.307 |
0.102 |
| 97 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.301 |
0.446 |
| 98 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.296 |
0.697 |
| 99 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.296 |
-0.252 |
| 100 |
253814_at |
AT4G28290
|
unknown |
-0.295 |
0.548 |
| 101 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
-0.290 |
-0.028 |
| 102 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.287 |
-0.036 |
| 103 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
-0.283 |
0.422 |
| 104 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.279 |
0.317 |
| 105 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.275 |
0.143 |
| 106 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.275 |
0.397 |
| 107 |
249798_at |
AT5G23730
|
EFO2, EARLY FLOWERING BY OVEREXPRESSION 2, RUP2, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 |
-0.273 |
0.660 |
| 108 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
-0.271 |
0.165 |
| 109 |
247452_at |
AT5G62430
|
CDF1, cycling DOF factor 1 |
-0.271 |
0.065 |
| 110 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.271 |
0.545 |
| 111 |
247933_at |
AT5G56980
|
unknown |
-0.269 |
0.524 |
| 112 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
-0.268 |
0.424 |
| 113 |
246142_at |
AT5G19970
|
unknown |
-0.265 |
0.064 |
| 114 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.264 |
0.613 |
| 115 |
255627_at |
AT4G00955
|
unknown |
-0.261 |
0.209 |
| 116 |
254544_at |
AT4G19820
|
[Glycosyl hydrolase family protein with chitinase insertion domain] |
-0.259 |
0.318 |
| 117 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.256 |
0.429 |
| 118 |
249932_at |
AT5G22390
|
unknown |
-0.256 |
0.273 |