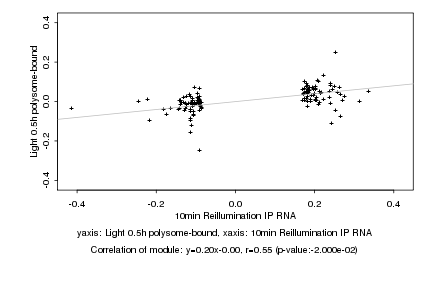
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
10min Reillumination IP RNA |
Log2 signal ratio
Light 0.5h polysome-bound |
| 1 |
262952_at |
AT1G75770
|
unknown |
0.336 |
0.055 |
| 2 |
255232_at |
AT4G05330
|
AGD13, ARF-GAP domain 13 |
0.311 |
0.003 |
| 3 |
251554_at |
AT3G58670
|
unknown |
0.275 |
0.030 |
| 4 |
260273_at |
AT1G80550
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.269 |
0.009 |
| 5 |
250124_at |
AT5G16480
|
AtPFA-DSP5, PFA-DSP5, plant and fungi atypical dual-specificity phosphatase 5 |
0.264 |
0.040 |
| 6 |
254362_at |
AT4G22160
|
unknown |
0.264 |
-0.075 |
| 7 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.262 |
0.075 |
| 8 |
265394_at |
AT2G20725
|
[CAAX amino terminal protease family protein] |
0.256 |
0.050 |
| 9 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.251 |
0.253 |
| 10 |
266811_at |
AT2G44850
|
unknown |
0.251 |
-0.045 |
| 11 |
258135_at |
AT3G24506
|
unknown |
0.248 |
0.079 |
| 12 |
250310_at |
AT5G12230
|
MED19A |
0.244 |
0.065 |
| 13 |
248509_at |
AT5G50335
|
unknown |
0.241 |
-0.109 |
| 14 |
261317_at |
AT1G53030
|
[Cytochrome C oxidase copper chaperone (COX17)] |
0.240 |
0.092 |
| 15 |
261941_at |
AT1G22490
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.240 |
-0.008 |
| 16 |
254626_at |
AT4G18400
|
unknown |
0.239 |
0.086 |
| 17 |
247833_at |
AT5G58575
|
unknown |
0.237 |
0.054 |
| 18 |
250972_at |
AT5G02840
|
LCL1, LHY/CCA1-like 1 |
0.236 |
0.023 |
| 19 |
245261_at |
AT4G14385
|
unknown |
0.222 |
0.136 |
| 20 |
250755_at |
AT5G05750
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.221 |
0.011 |
| 21 |
258092_at |
AT3G14595
|
[Ribosomal protein L18ae family] |
0.216 |
0.048 |
| 22 |
255931_at |
AT1G12710
|
AtPP2-A12, phloem protein 2-A12, PP2-A12, phloem protein 2-A12 |
0.214 |
0.044 |
| 23 |
255506_at |
AT4G02130
|
GATL6, galacturonosyltransferase-like 6, LGT10 |
0.211 |
-0.001 |
| 24 |
250555_at |
AT5G07950
|
unknown |
0.210 |
0.054 |
| 25 |
246951_at |
AT5G04880
|
[expressed protein] |
0.208 |
-0.013 |
| 26 |
245797_at |
AT1G45230
|
unknown |
0.208 |
0.102 |
| 27 |
245778_at |
AT1G73530
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.205 |
0.107 |
| 28 |
248869_at |
AT5G46840
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.204 |
0.010 |
| 29 |
266238_at |
AT2G29400
|
PP1-AT, PROTEIN PHOSPHATASE 1, TOPP1, type one protein phosphatase 1 |
0.204 |
0.021 |
| 30 |
249872_at |
AT5G23130
|
[Peptidoglycan-binding LysM domain-containing protein] |
0.202 |
0.015 |
| 31 |
249990_at |
AT5G18540
|
unknown |
0.202 |
0.006 |
| 32 |
265142_at |
AT1G51360
|
ATDABB1, DIMERIC A/B BARREL DOMAINS-PROTEIN 1, DABB1, dimeric A/B barrel domainS-protein 1 |
0.202 |
0.081 |
| 33 |
257018_at |
AT3G19630
|
[Radical SAM superfamily protein] |
0.200 |
0.062 |
| 34 |
260964_at |
AT1G45050
|
ATUBC2-1, UBC15, Arabidopsis thaliana ubiquitin-conjugating enzyme 15 |
0.200 |
0.069 |
| 35 |
258126_at |
AT3G24490
|
[Alcohol dehydrogenase transcription factor Myb/SANT-like family protein] |
0.199 |
0.041 |
| 36 |
250856_at |
AT5G04810
|
[pentatricopeptide (PPR) repeat-containing protein] |
0.196 |
0.031 |
| 37 |
250017_at |
AT5G18140
|
DJC69, DNA J protein A69 |
0.195 |
0.071 |
| 38 |
263839_at |
AT2G36900
|
ATMEMB11, MEMB11, membrin 11 |
0.195 |
0.064 |
| 39 |
249524_at |
AT5G38520
|
[alpha/beta-Hydrolases superfamily protein] |
0.194 |
0.073 |
| 40 |
246319_at |
AT3G56680
|
[Single-stranded nucleic acid binding R3H protein] |
0.191 |
0.034 |
| 41 |
246506_at |
AT5G16110
|
unknown |
0.189 |
0.001 |
| 42 |
250175_at |
AT5G14390
|
[alpha/beta-Hydrolases superfamily protein] |
0.188 |
0.001 |
| 43 |
249966_at |
AT5G19030
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.188 |
0.012 |
| 44 |
262498_at |
AT1G21710
|
ATOGG1, ARABIDOPSIS 8-OXOGUANINE-DNA GLYCOSYLASE 1, OGG1, 8-oxoguanine-DNA glycosylase 1 |
0.187 |
0.058 |
| 45 |
267405_at |
AT2G33740
|
CUTA |
0.186 |
0.076 |
| 46 |
261823_at |
AT1G11400
|
PYM, partner of Y14-MAGO |
0.184 |
0.062 |
| 47 |
253484_at |
AT4G31720
|
STG1, SALT TOLERANCE DURING GERMINATION 1, TAF10, TBP-ASSOCIATED FACTOR 10, TAFII15, TBP-associated factor II 15 |
0.183 |
0.049 |
| 48 |
256133_at |
AT1G13570
|
[F-box/RNI-like superfamily protein] |
0.182 |
0.042 |
| 49 |
250567_at |
AT5G08090
|
unknown |
0.182 |
0.030 |
| 50 |
254477_at |
AT4G20380
|
LSD1, LESION SIMULATING DISEASE 1 |
0.181 |
0.046 |
| 51 |
255878_at |
AT2G40620
|
[Basic-leucine zipper (bZIP) transcription factor family protein] |
0.181 |
-0.024 |
| 52 |
266003_at |
AT2G37320
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.181 |
0.004 |
| 53 |
249544_at |
AT5G38110
|
ASF1B, anti- silencing function 1b, SGA01, SGA1 |
0.181 |
0.083 |
| 54 |
266051_at |
AT2G40760
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.180 |
0.061 |
| 55 |
267375_at |
AT2G26300
|
ATGPA1, ARABIDOPSIS THALIANA G PROTEIN ALPHA SUBUNIT 1, GP ALPHA 1, G protein alpha subunit 1, GPA1, G PROTEIN ALPHA SUBUNIT 1 |
0.180 |
0.053 |
| 56 |
250896_at |
AT5G03560
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.178 |
0.092 |
| 57 |
264265_at |
AT1G09280
|
unknown |
0.178 |
0.018 |
| 58 |
263288_at |
AT2G36130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.177 |
0.051 |
| 59 |
266923_at |
AT2G45980
|
ATI1, ATG8-interacting protein 1 |
0.177 |
0.080 |
| 60 |
252918_at |
AT4G38980
|
unknown |
0.174 |
0.068 |
| 61 |
247908_at |
AT5G57440
|
GPP2, GLYCEROL-3-PHOSPHATASE 2, GS1 |
0.174 |
0.008 |
| 62 |
257818_at |
AT3G25120
|
[Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein] |
0.173 |
0.046 |
| 63 |
261084_at |
AT1G07440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.172 |
0.104 |
| 64 |
261757_at |
AT1G08210
|
[Eukaryotic aspartyl protease family protein] |
0.172 |
0.019 |
| 65 |
250837_at |
AT5G04620
|
ATBIOF, biotin F, BIO4, biotin 4, BIOF, biotin F |
0.171 |
0.041 |
| 66 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.170 |
0.039 |
| 67 |
259378_at |
AT3G16310
|
1 |
0.170 |
0.038 |
| 68 |
261272_at |
AT1G26665
|
[Mediator complex, subunit Med10] |
0.169 |
0.062 |
| 69 |
264586_at |
AT1G05160
|
ATKAO1, CYP88A3, cytochrome P450, family 88, subfamily A, polypeptide 3, KAO1, ENT-KAURENOIC ACID OXYDASE 1 |
0.169 |
0.009 |
| 70 |
244973_at |
ATCG00690
|
PSBT, photosystem II reaction center protein T, PSBTC |
-0.416 |
-0.034 |
| 71 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
-0.247 |
0.002 |
| 72 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.224 |
0.015 |
| 73 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.218 |
-0.096 |
| 74 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.184 |
-0.037 |
| 75 |
263221_at |
AT1G30620
|
HSR8, HIGH SUGAR RESPONSE8, MUR4, MURUS 4, UXE1, UDP-D-XYLOSE 4-EPIMERASE 1 |
-0.176 |
-0.061 |
| 76 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
-0.165 |
-0.033 |
| 77 |
260987_at |
AT1G53590
|
NTMC2T6.1, NTMC2TYPE6.1 |
-0.144 |
-0.038 |
| 78 |
250161_at |
AT5G15240
|
[Transmembrane amino acid transporter family protein] |
-0.143 |
-0.032 |
| 79 |
258601_at |
AT3G02760
|
[Class II aaRS and biotin synthetases superfamily protein] |
-0.142 |
0.009 |
| 80 |
257846_at |
AT3G12910
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
-0.140 |
0.009 |
| 81 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
-0.139 |
0.007 |
| 82 |
256952_at |
AT3G21160
|
MANIA, ALPHA-MANNOSIDASE IA, MNS2, alpha-mannosidase 2 |
-0.139 |
-0.013 |
| 83 |
255259_at |
AT4G05020
|
NDB2, NAD(P)H dehydrogenase B2 |
-0.138 |
-0.000 |
| 84 |
263219_at |
AT1G30600
|
[Subtilase family protein] |
-0.133 |
0.023 |
| 85 |
246500_at |
AT5G16270
|
ATRAD21.3, ARABIDOPSIS HOMOLOG OF RAD21 3, SYN4, sister chromatid cohesion 1 protein 4 |
-0.132 |
-0.002 |
| 86 |
262766_at |
AT1G13160
|
[ARM repeat superfamily protein] |
-0.131 |
-0.044 |
| 87 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.128 |
-0.565 |
| 88 |
244962_at |
ATCG01050
|
NDHD |
-0.128 |
-0.010 |
| 89 |
254847_at |
AT4G11850
|
MEE54, maternal effect embryo arrest 54, PLDGAMMA1, phospholipase D gamma 1 |
-0.128 |
-0.033 |
| 90 |
263130_at |
AT1G78650
|
POLD3 |
-0.128 |
-0.010 |
| 91 |
244998_at |
ATCG00180
|
RPOC1 |
-0.126 |
-0.021 |
| 92 |
264932_at |
AT1G61240
|
unknown |
-0.126 |
0.027 |
| 93 |
245047_at |
ATCG00020
|
PSBA, photosystem II reaction center protein A |
-0.124 |
-0.014 |
| 94 |
251752_at |
AT3G55740
|
ATPROT2, PROLINE TRANSPORTER 2, PROT2, proline transporter 2 |
-0.119 |
-0.010 |
| 95 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.117 |
0.037 |
| 96 |
260143_at |
AT1G71880
|
ATSUC1, ARABIDOPSIS THALIANA SUCROSE-PROTON SYMPORTER 1, SUC1, sucrose-proton symporter 1 |
-0.116 |
-0.092 |
| 97 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
-0.115 |
0.026 |
| 98 |
247153_at |
AT5G65700
|
BAM1, BARELY ANY MERISTEM 1 |
-0.115 |
-0.036 |
| 99 |
250021_at |
AT5G18190
|
[Protein kinase family protein] |
-0.114 |
-0.010 |
| 100 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.114 |
-0.049 |
| 101 |
258179_at |
AT3G21690
|
[MATE efflux family protein] |
-0.114 |
-0.037 |
| 102 |
259015_at |
AT3G07350
|
unknown |
-0.114 |
-0.156 |
| 103 |
251916_at |
AT3G53960
|
[Major facilitator superfamily protein] |
-0.114 |
-0.085 |
| 104 |
259982_at |
AT1G76410
|
ATL8 |
-0.112 |
-0.121 |
| 105 |
264330_at |
AT1G04120
|
ABCC5, ATP-binding cassette C5, ATABCC5, Arabidopsis thaliana ATP-binding cassette C5, ATMRP5, multidrug resistance-associated protein 5, MRP5, MULTIDRUG RESISTANCE PROTEIN 5, MRP5, multidrug resistance-associated protein 5 |
-0.112 |
-0.002 |
| 106 |
250490_at |
AT5G09760
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.111 |
-0.014 |
| 107 |
260374_at |
AT1G73960
|
TAF2, TBP-associated factor 2 |
-0.111 |
-0.005 |
| 108 |
252149_at |
AT3G51290
|
APSR1, Altered Phosphate Starvation Response 1 |
-0.109 |
-0.024 |
| 109 |
254922_at |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
-0.109 |
0.020 |
| 110 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
-0.109 |
0.010 |
| 111 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.108 |
-0.069 |
| 112 |
250787_at |
AT5G05560
|
APC1, anaphase promoting complex 1, EMB2771, EMBRYO DEFECTIVE 2771 |
-0.107 |
-0.049 |
| 113 |
247425_at |
AT5G62550
|
unknown |
-0.106 |
-0.065 |
| 114 |
258555_at |
AT3G06860
|
ATMFP2, MULTIFUNCTIONAL PROTEIN 2, MFP2, multifunctional protein 2 |
-0.105 |
-0.012 |
| 115 |
266299_at |
AT2G29450
|
AT103-1A, ATGSTU1, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 1, ATGSTU5, glutathione S-transferase tau 5, GSTU5, glutathione S-transferase tau 5 |
-0.104 |
0.075 |
| 116 |
246660_at |
AT5G35180
|
[FUNCTIONS IN: phosphoinositide binding] |
-0.104 |
-0.000 |
| 117 |
248615_at |
AT5G49570
|
AtPNG1, peptide-N-glycanase 1, PNG1, peptide-N-glycanase 1 |
-0.100 |
-0.005 |
| 118 |
260629_at |
AT1G62330
|
[O-fucosyltransferase family protein] |
-0.097 |
0.006 |
| 119 |
254904_at |
AT4G11160
|
[Translation initiation factor 2, small GTP-binding protein] |
-0.097 |
0.021 |
| 120 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.096 |
0.041 |
| 121 |
267376_at |
AT2G26330
|
ER, ERECTA, QRP1, QUANTITATIVE RESISTANCE TO PLECTOSPHAERELLA 1 |
-0.094 |
-0.003 |
| 122 |
250077_at |
AT5G16680
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.094 |
0.019 |
| 123 |
255460_at |
AT4G02800
|
unknown |
-0.093 |
-0.044 |
| 124 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
-0.093 |
-0.011 |
| 125 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.092 |
-0.246 |
| 126 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
-0.091 |
0.028 |
| 127 |
255039_at |
AT4G09570
|
ATCPK4, CPK4, calcium-dependent protein kinase 4 |
-0.091 |
0.014 |
| 128 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
-0.091 |
0.067 |
| 129 |
267618_at |
AT2G26760
|
CYCB1;4, Cyclin B1;4 |
-0.091 |
0.006 |
| 130 |
266017_at |
AT2G18690
|
unknown |
-0.090 |
-0.003 |
| 131 |
246763_at |
AT5G27150
|
AT-NHX1, ATNHX, ATNHX1, NHX1, Na+/H+ exchanger 1 |
-0.090 |
-0.004 |
| 132 |
260944_at |
AT1G45130
|
AtBGAL5, BGAL5, beta-galactosidase 5 |
-0.090 |
-0.018 |
| 133 |
245050_at |
ATCG00070
|
PSBK, photosystem II reaction center protein K precursor |
-0.089 |
-0.002 |
| 134 |
264352_at |
AT1G03270
|
unknown |
-0.089 |
-0.024 |
| 135 |
256908_at |
AT3G24040
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.088 |
-0.001 |
| 136 |
249095_at |
AT5G43900
|
ATMYA2, ARABIDOPSIS MYOSIN 2, MYA2, myosin 2, XI-2, MYOSIN X1 2, XI-6, MYOSIN XI-6 |
-0.088 |
-0.035 |
| 137 |
258132_at |
AT3G24550
|
ATPERK1, proline-rich extensin-like receptor kinase 1, PERK1, proline-rich extensin-like receptor kinase 1 |
-0.088 |
-0.027 |