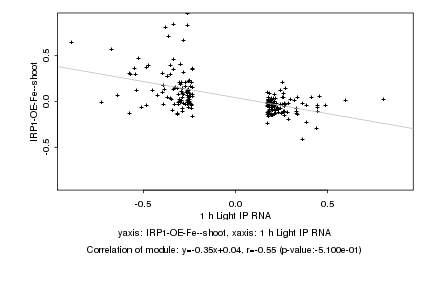
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
1 h Light IP RNA |
Log2 signal ratio
IRP1-OE-Fe--shoot |
| 1 |
252661_at |
AT3G44450
|
unknown |
0.802 |
0.031 |
| 2 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.595 |
0.021 |
| 3 |
252010_at |
AT3G52740
|
unknown |
0.486 |
-0.041 |
| 4 |
261247_at |
AT1G20070
|
unknown |
0.454 |
0.056 |
| 5 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
0.445 |
-0.064 |
| 6 |
245341_at |
AT4G16447
|
unknown |
0.444 |
-0.098 |
| 7 |
266760_at |
AT2G46870
|
NGA1, NGATHA1 |
0.444 |
-0.043 |
| 8 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
0.439 |
-0.287 |
| 9 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.413 |
0.051 |
| 10 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
0.383 |
-0.225 |
| 11 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
0.381 |
-0.043 |
| 12 |
263715_at |
AT2G20570
|
GBF's pro-rich region-interacting factor 1 |
0.363 |
-0.019 |
| 13 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
0.363 |
-0.408 |
| 14 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
0.335 |
0.047 |
| 15 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.334 |
-0.131 |
| 16 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.331 |
-0.072 |
| 17 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
0.331 |
-0.130 |
| 18 |
248097_at |
AT5G55260
|
PPX-2, PROTEIN PHOSPHATASE X -2, PPX2, protein phosphatase X 2 |
0.329 |
-0.104 |
| 19 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
0.319 |
0.016 |
| 20 |
246313_at |
AT1G31920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.297 |
0.024 |
| 21 |
262421_at |
AT1G50290
|
unknown |
0.284 |
-0.190 |
| 22 |
267294_at |
AT2G23670
|
YCF37, homolog of Synechocystis YCF37 |
0.283 |
-0.017 |
| 23 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
0.278 |
-0.114 |
| 24 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
0.271 |
0.146 |
| 25 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.271 |
-0.032 |
| 26 |
257954_at |
AT3G21760
|
HYR1, HYPOSTATIN RESISTANCE 1 |
0.266 |
-0.106 |
| 27 |
262162_at |
AT1G78020
|
unknown |
0.264 |
-0.020 |
| 28 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
0.264 |
-0.145 |
| 29 |
260354_at |
AT1G69330
|
[RING/U-box superfamily protein] |
0.262 |
-0.050 |
| 30 |
260696_at |
AT1G32520
|
unknown |
0.261 |
-0.115 |
| 31 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
0.261 |
0.088 |
| 32 |
264057_at |
AT2G28550
|
RAP2.7, related to AP2.7, TOE1, TARGET OF EARLY ACTIVATION TAGGED (EAT) 1 |
0.260 |
-0.105 |
| 33 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
0.258 |
0.044 |
| 34 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
0.255 |
0.209 |
| 35 |
258587_at |
AT3G04310
|
unknown |
0.254 |
0.049 |
| 36 |
246986_at |
AT5G67280
|
RLK, receptor-like kinase |
0.251 |
-0.026 |
| 37 |
256819_at |
AT3G21390
|
[Mitochondrial substrate carrier family protein] |
0.245 |
-0.123 |
| 38 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.244 |
0.123 |
| 39 |
255506_at |
AT4G02130
|
GATL6, galacturonosyltransferase-like 6, LGT10 |
0.244 |
-0.074 |
| 40 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
0.243 |
-0.022 |
| 41 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
0.234 |
-0.079 |
| 42 |
256049_at |
AT1G07010
|
AtSLP1, SLP1, Shewenella-like protein phosphatase 1 |
0.233 |
-0.113 |
| 43 |
249107_at |
AT5G43680
|
unknown |
0.227 |
-0.084 |
| 44 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
0.224 |
-0.056 |
| 45 |
258621_at |
AT3G02830
|
PNT1, PENTA 1, ZFN1, zinc finger protein 1 |
0.223 |
-0.018 |
| 46 |
250981_at |
AT5G03140
|
LecRK-VIII.2, L-type lectin receptor kinase VIII.2 |
0.223 |
-0.082 |
| 47 |
248291_at |
AT5G53020
|
[Ribonuclease P protein subunit P38-related] |
0.215 |
-0.096 |
| 48 |
258720_at |
AT3G09550
|
[Ankyrin repeat family protein] |
0.214 |
-0.129 |
| 49 |
254693_at |
AT4G17880
|
MYC4 |
0.214 |
0.042 |
| 50 |
264206_at |
AT1G22730
|
[MA3 domain-containing protein] |
0.212 |
0.077 |
| 51 |
246319_at |
AT3G56680
|
[Single-stranded nucleic acid binding R3H protein] |
0.212 |
-0.047 |
| 52 |
250926_at |
AT5G03555
|
AtNCS1, NCS1, nucleobase cation symporter 1 |
0.211 |
-0.125 |
| 53 |
262938_at |
AT1G79540
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.211 |
0.009 |
| 54 |
264587_at |
AT1G05200
|
ATGLR3.4, glutamate receptor 3.4, GLR3.4, glutamate receptor 3.4, GLUR3 |
0.210 |
-0.136 |
| 55 |
250721_at |
AT5G06210
|
[RNA binding (RRM/RBD/RNP motifs) family protein] |
0.209 |
-0.021 |
| 56 |
247141_at |
AT5G65560
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.209 |
-0.081 |
| 57 |
245152_at |
AT2G47490
|
ATNDT1, ARABIDOPSIS THALIANA NAD+ TRANSPORTER 1, NDT1, NAD+ transporter 1 |
0.207 |
-0.038 |
| 58 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
0.205 |
0.035 |
| 59 |
249798_at |
AT5G23730
|
EFO2, EARLY FLOWERING BY OVEREXPRESSION 2, RUP2, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 |
0.204 |
-0.012 |
| 60 |
261898_at |
AT1G80720
|
[Mitochondrial glycoprotein family protein] |
0.202 |
-0.014 |
| 61 |
262711_at |
AT1G16500
|
unknown |
0.199 |
-0.078 |
| 62 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
0.199 |
-0.041 |
| 63 |
266164_at |
AT2G28050
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.199 |
-0.097 |
| 64 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
0.198 |
-0.063 |
| 65 |
255825_at |
AT2G40475
|
ASG8, ALTERED SEED GERMINATION 8 |
0.197 |
-0.144 |
| 66 |
260922_at |
AT1G21560
|
unknown |
0.196 |
-0.143 |
| 67 |
251263_at |
AT3G62190
|
[Chaperone DnaJ-domain superfamily protein] |
0.195 |
0.019 |
| 68 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
0.194 |
-0.004 |
| 69 |
249524_at |
AT5G38520
|
[alpha/beta-Hydrolases superfamily protein] |
0.194 |
-0.142 |
| 70 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
0.193 |
-0.079 |
| 71 |
255130_at |
AT4G08240
|
unknown |
0.193 |
-0.015 |
| 72 |
253545_at |
AT4G31310
|
[AIG2-like (avirulence induced gene) family protein] |
0.192 |
-0.041 |
| 73 |
264920_at |
AT1G60550
|
DHNS, ECHID, enoyl-CoA hydratase/isomerase D |
0.192 |
0.033 |
| 74 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
0.191 |
-0.139 |
| 75 |
253440_at |
AT4G32570
|
TIFY8, TIFY domain protein 8 |
0.191 |
0.023 |
| 76 |
247966_at |
AT5G56610
|
[Phosphotyrosine protein phosphatases superfamily protein] |
0.187 |
-0.008 |
| 77 |
250075_at |
AT5G17670
|
[alpha/beta-Hydrolases superfamily protein] |
0.186 |
-0.141 |
| 78 |
260964_at |
AT1G45050
|
ATUBC2-1, UBC15, Arabidopsis thaliana ubiquitin-conjugating enzyme 15 |
0.185 |
-0.034 |
| 79 |
248693_at |
AT5G48330
|
RUG2, RCC1/UVR8/GEF-like 2 |
0.185 |
-0.098 |
| 80 |
259287_at |
AT3G11490
|
[rac GTPase activating protein] |
0.184 |
-0.065 |
| 81 |
267504_at |
AT2G45530
|
[RING/U-box superfamily protein] |
0.183 |
0.007 |
| 82 |
245778_at |
AT1G73530
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.183 |
-0.081 |
| 83 |
245612_at |
AT4G14440
|
ATECI3, ARABIDOPSIS THALIANA DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, ECI3, DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, HCD1, 3-hydroxyacyl-CoA dehydratase 1 |
0.182 |
-0.041 |
| 84 |
259378_at |
AT3G16310
|
1 |
0.182 |
-0.010 |
| 85 |
267505_at |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
0.181 |
0.092 |
| 86 |
261813_at |
AT1G08280
|
[Glycosyltransferase family 29 (sialyltransferase) family protein] |
0.181 |
-0.004 |
| 87 |
249829_at |
AT5G23290
|
PFD5, prefoldin 5 |
0.178 |
-0.021 |
| 88 |
262464_at |
AT1G50280
|
[Phototropic-responsive NPH3 family protein] |
0.178 |
-0.101 |
| 89 |
248028_at |
AT5G55620
|
unknown |
0.178 |
-0.054 |
| 90 |
259284_at |
AT3G11450
|
[DnaJ domain ] |
0.177 |
0.012 |
| 91 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.177 |
0.031 |
| 92 |
265742_at |
AT2G01290
|
RPI2, ribose-5-phosphate isomerase 2 |
0.176 |
-0.028 |
| 93 |
264583_at |
AT1G05170
|
[Galactosyltransferase family protein] |
0.176 |
0.054 |
| 94 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
0.176 |
-0.159 |
| 95 |
263236_at |
AT1G10470
|
ARR4, response regulator 4, ATRR1, RESPONCE REGULATOR 1, IBC7, INDUCED BY CYTOKININ 7, MEE7, maternal effect embryo arrest 7 |
0.176 |
0.004 |
| 96 |
246015_at |
AT5G10700
|
[Peptidyl-tRNA hydrolase II (PTH2) family protein] |
0.176 |
-0.121 |
| 97 |
260273_at |
AT1G80550
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.175 |
0.032 |
| 98 |
264440_at |
AT1G27340
|
LCR, LEAF CURLING RESPONSIVENESS |
0.175 |
0.019 |
| 99 |
260961_at |
AT1G05960
|
[ARM repeat superfamily protein] |
0.173 |
-0.035 |
| 100 |
264738_at |
AT1G62250
|
unknown |
0.173 |
-0.132 |
| 101 |
261772_at |
AT1G76240
|
unknown |
0.173 |
0.109 |
| 102 |
256071_at |
AT1G13640
|
[Phosphatidylinositol 3- and 4-kinase family protein] |
0.172 |
-0.119 |
| 103 |
245682_at |
AT5G08750
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.171 |
-0.015 |
| 104 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
0.171 |
-0.047 |
| 105 |
258726_at |
AT3G11745
|
unknown |
0.171 |
-0.239 |
| 106 |
266197_at |
AT2G39120
|
WTF9, what's this factor 9 |
0.170 |
-0.008 |
| 107 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
-0.895 |
0.652 |
| 108 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-0.733 |
-0.004 |
| 109 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
-0.677 |
0.578 |
| 110 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
-0.643 |
0.068 |
| 111 |
247324_at |
AT5G64190
|
unknown |
-0.580 |
0.311 |
| 112 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
-0.580 |
-0.121 |
| 113 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
-0.571 |
0.304 |
| 114 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-0.554 |
0.368 |
| 115 |
244973_at |
ATCG00690
|
PSBT, photosystem II reaction center protein T, PSBTC |
-0.544 |
0.305 |
| 116 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-0.543 |
0.127 |
| 117 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
-0.527 |
0.480 |
| 118 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.511 |
-0.062 |
| 119 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
-0.487 |
0.372 |
| 120 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
-0.485 |
-0.040 |
| 121 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-0.474 |
0.403 |
| 122 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
-0.455 |
0.124 |
| 123 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.427 |
0.070 |
| 124 |
245007_at |
ATCG00350
|
PSAA |
-0.397 |
0.100 |
| 125 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-0.397 |
0.309 |
| 126 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
-0.395 |
-0.025 |
| 127 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
-0.394 |
0.176 |
| 128 |
247137_at |
AT5G66210
|
CPK28, calcium-dependent protein kinase 28 |
-0.387 |
0.135 |
| 129 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
-0.381 |
0.815 |
| 130 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.372 |
0.052 |
| 131 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.371 |
0.274 |
| 132 |
266140_at |
AT2G28120
|
[Major facilitator superfamily protein] |
-0.370 |
0.279 |
| 133 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.368 |
0.718 |
| 134 |
244972_at |
ATCG00680
|
PSBB, photosystem II reaction center protein B |
-0.358 |
0.401 |
| 135 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.358 |
0.040 |
| 136 |
259982_at |
AT1G76410
|
ATL8 |
-0.357 |
0.303 |
| 137 |
253864_at |
AT4G27460
|
CBSX5, CBS domain containing protein 5 |
-0.348 |
0.025 |
| 138 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.343 |
-0.089 |
| 139 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
-0.341 |
0.465 |
| 140 |
253322_at |
AT4G33980
|
unknown |
-0.340 |
0.138 |
| 141 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
-0.338 |
0.845 |
| 142 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.337 |
0.359 |
| 143 |
260969_at |
AT1G12240
|
ATBETAFRUCT4, AtFRUCT4, AtVI2, FRUCT4, fructosidase 4, VAC-INV, VACUOLAR INVERTASE, VI2, vacuolar invertase 2 |
-0.335 |
0.147 |
| 144 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.332 |
-0.027 |
| 145 |
265732_at |
AT2G01300
|
unknown |
-0.329 |
0.155 |
| 146 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
-0.317 |
-0.124 |
| 147 |
246018_at |
AT5G10695
|
unknown |
-0.317 |
0.149 |
| 148 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
-0.316 |
-0.138 |
| 149 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
-0.314 |
-0.022 |
| 150 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
-0.311 |
0.079 |
| 151 |
264617_at |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
-0.311 |
-0.006 |
| 152 |
262525_at |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
-0.308 |
0.007 |
| 153 |
256799_at |
AT3G18560
|
unknown |
-0.307 |
0.208 |
| 154 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
-0.307 |
0.025 |
| 155 |
264867_at |
AT1G24150
|
ATFH4, FORMIN HOMOLOGUE 4, FH4, formin homologue 4 |
-0.304 |
0.188 |
| 156 |
252991_at |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
-0.303 |
-0.004 |
| 157 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
-0.300 |
0.408 |
| 158 |
261567_at |
AT1G33055
|
unknown |
-0.299 |
0.411 |
| 159 |
260411_at |
AT1G69890
|
unknown |
-0.298 |
0.014 |
| 160 |
253819_at |
AT4G28350
|
LecRK-VII.2, L-type lectin receptor kinase VII.2 |
-0.297 |
0.120 |
| 161 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-0.296 |
0.208 |
| 162 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
-0.290 |
0.147 |
| 163 |
263118_at |
AT1G03090
|
MCCA |
-0.290 |
-0.067 |
| 164 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
-0.289 |
-0.107 |
| 165 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.289 |
0.047 |
| 166 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
-0.288 |
0.089 |
| 167 |
248502_at |
AT5G50450
|
[HCP-like superfamily protein with MYND-type zinc finger] |
-0.284 |
0.082 |
| 168 |
245353_at |
AT4G16000
|
unknown |
-0.284 |
0.323 |
| 169 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
-0.283 |
-0.011 |
| 170 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.283 |
0.667 |
| 171 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
-0.274 |
0.174 |
| 172 |
260126_at |
AT1G36370
|
SHM7, serine hydroxymethyltransferase 7 |
-0.273 |
-0.029 |
| 173 |
245018_at |
ATCG00520
|
YCF4 |
-0.273 |
0.209 |
| 174 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
-0.269 |
0.103 |
| 175 |
253284_at |
AT4G34150
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.266 |
0.124 |
| 176 |
259792_at |
AT1G29690
|
CAD1, constitutively activated cell death 1 |
-0.266 |
0.017 |
| 177 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
-0.265 |
0.831 |
| 178 |
257076_at |
AT3G19680
|
unknown |
-0.265 |
-0.002 |
| 179 |
262456_at |
AT1G11260
|
ATSTP1, SUGAR TRANSPORTER 1, STP1, sugar transporter 1 |
-0.264 |
0.114 |
| 180 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
-0.263 |
0.963 |
| 181 |
259479_at |
AT1G19020
|
unknown |
-0.262 |
0.102 |
| 182 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
-0.261 |
0.076 |
| 183 |
254948_at |
AT4G11000
|
[Ankyrin repeat family protein] |
-0.261 |
-0.027 |
| 184 |
262026_at |
AT1G35670
|
ATCDPK2, calcium-dependent protein kinase 2, ATCPK11, CDPK2, calcium-dependent protein kinase 2, CPK11 |
-0.260 |
0.010 |
| 185 |
267393_at |
AT2G44500
|
[O-fucosyltransferase family protein] |
-0.260 |
0.109 |
| 186 |
255872_at |
AT2G30360
|
CIPK11, CBL-INTERACTING PROTEIN KINASE 11, PKS5, PROTEIN KINASE SOS2-LIKE 5, SIP4, SOS3-interacting protein 4, SNRK3.22, SNF1-RELATED PROTEIN KINASE 3.22 |
-0.260 |
0.140 |
| 187 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
-0.259 |
-0.005 |
| 188 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
-0.259 |
0.224 |
| 189 |
255039_at |
AT4G09570
|
ATCPK4, CPK4, calcium-dependent protein kinase 4 |
-0.259 |
0.029 |
| 190 |
248123_at |
AT5G54720
|
[Ankyrin repeat family protein] |
-0.257 |
0.048 |
| 191 |
264836_at |
AT1G03610
|
unknown |
-0.256 |
-0.058 |
| 192 |
264652_at |
AT1G08920
|
ESL1, ERD (early response to dehydration) six-like 1 |
-0.256 |
0.127 |
| 193 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
-0.255 |
0.011 |
| 194 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.253 |
0.065 |
| 195 |
259871_at |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
-0.252 |
0.026 |
| 196 |
263478_at |
AT2G31880
|
EVR, EVERSHED, SOBIR1, SUPPRESSOR OF BIR1 1 |
-0.251 |
0.098 |
| 197 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
-0.250 |
0.236 |
| 198 |
252483_at |
AT3G46600
|
[GRAS family transcription factor] |
-0.247 |
-0.019 |
| 199 |
261052_at |
AT1G01440
|
unknown |
-0.247 |
0.062 |
| 200 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
-0.245 |
0.169 |
| 201 |
246584_at |
AT5G14730
|
unknown |
-0.244 |
-0.039 |
| 202 |
245026_at |
ATCG00140
|
ATPH |
-0.242 |
0.211 |
| 203 |
250809_at |
AT5G05140
|
[Transcription elongation factor (TFIIS) family protein] |
-0.241 |
0.211 |
| 204 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
-0.240 |
-0.069 |
| 205 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
-0.240 |
-0.024 |
| 206 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
-0.238 |
0.364 |
| 207 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
-0.238 |
0.359 |
| 208 |
261822_at |
AT1G11380
|
[PLAC8 family protein] |
-0.238 |
-0.162 |
| 209 |
259015_at |
AT3G07350
|
unknown |
-0.237 |
0.058 |
| 210 |
262452_at |
AT1G11210
|
unknown |
-0.236 |
0.082 |
| 211 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
-0.236 |
0.157 |