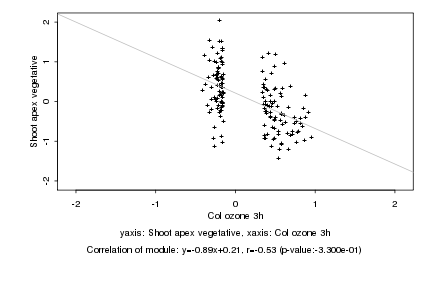
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col ozone 3h |
Log2 signal ratio
Shoot apex vegetative |
| 1 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.949 |
-0.893 |
| 2 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.909 |
-0.273 |
| 3 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.877 |
-0.400 |
| 4 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.867 |
0.169 |
| 5 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.863 |
-0.967 |
| 6 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.841 |
-0.156 |
| 7 |
264774_at |
AT1G22890
|
unknown |
0.829 |
-0.628 |
| 8 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.814 |
-0.539 |
| 9 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.812 |
-0.419 |
| 10 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.786 |
-0.748 |
| 11 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
0.777 |
-0.759 |
| 12 |
261394_at |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
0.764 |
-0.488 |
| 13 |
252346_at |
AT3G48650
|
[pseudogene] |
0.755 |
-1.030 |
| 14 |
257846_at |
AT3G12910
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
0.732 |
-0.393 |
| 15 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.731 |
-0.550 |
| 16 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.711 |
-0.809 |
| 17 |
254922_at |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
0.710 |
-0.736 |
| 18 |
259479_at |
AT1G19020
|
unknown |
0.687 |
0.391 |
| 19 |
254408_at |
AT4G21390
|
B120 |
0.681 |
-0.840 |
| 20 |
262085_at |
AT1G56060
|
unknown |
0.656 |
-0.143 |
| 21 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.655 |
-1.200 |
| 22 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.652 |
-0.783 |
| 23 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.616 |
-0.514 |
| 24 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.604 |
0.968 |
| 25 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.604 |
-0.329 |
| 26 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.588 |
0.344 |
| 27 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.578 |
-0.555 |
| 28 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
0.576 |
-0.302 |
| 29 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
0.576 |
0.149 |
| 30 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.575 |
-1.065 |
| 31 |
246370_at |
AT1G51920
|
unknown |
0.571 |
-1.041 |
| 32 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
0.567 |
-0.279 |
| 33 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
0.559 |
0.204 |
| 34 |
250267_at |
AT5G12930
|
unknown |
0.557 |
-0.464 |
| 35 |
255654_at |
AT4G00970
|
CRK41, cysteine-rich RLK (RECEPTOR-like protein kinase) 41 |
0.548 |
-1.201 |
| 36 |
264757_at |
AT1G61360
|
[S-locus lectin protein kinase family protein] |
0.538 |
-0.811 |
| 37 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.531 |
-0.731 |
| 38 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
0.528 |
-1.410 |
| 39 |
250796_at |
AT5G05300
|
unknown |
0.522 |
-0.108 |
| 40 |
263182_at |
AT1G05575
|
unknown |
0.499 |
0.336 |
| 41 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.496 |
1.188 |
| 42 |
266658_at |
AT2G25735
|
unknown |
0.492 |
-0.389 |
| 43 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.488 |
-0.469 |
| 44 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
0.488 |
0.893 |
| 45 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.488 |
-0.912 |
| 46 |
246993_at |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
0.481 |
-0.667 |
| 47 |
253044_at |
AT4G37290
|
unknown |
0.479 |
-0.428 |
| 48 |
251769_at |
AT3G55950
|
ATCRR3, CCR3, CRINKLY4 related 3 |
0.478 |
0.017 |
| 49 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.478 |
0.319 |
| 50 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.475 |
-0.952 |
| 51 |
254014_at |
AT4G26120
|
[Ankyrin repeat family protein / BTB/POZ domain-containing protein] |
0.460 |
-0.637 |
| 52 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.456 |
-0.041 |
| 53 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.446 |
-1.108 |
| 54 |
252133_at |
AT3G50900
|
unknown |
0.445 |
0.730 |
| 55 |
266017_at |
AT2G18690
|
unknown |
0.440 |
-0.108 |
| 56 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
0.438 |
-0.263 |
| 57 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.436 |
0.010 |
| 58 |
253779_at |
AT4G28490
|
HAE, HAESA, RLK5, RECEPTOR-LIKE PROTEIN KINASE 5 |
0.435 |
-0.382 |
| 59 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.430 |
-0.360 |
| 60 |
252053_at |
AT3G52400
|
ATSYP122, SYP122, syntaxin of plants 122 |
0.428 |
0.157 |
| 61 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.423 |
-0.116 |
| 62 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
0.404 |
1.217 |
| 63 |
259033_at |
AT3G09405
|
[Pectinacetylesterase family protein] |
0.400 |
0.296 |
| 64 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.395 |
-0.814 |
| 65 |
264663_at |
AT1G09970
|
LRR XI-23, RLK7, receptor-like kinase 7 |
0.395 |
-0.083 |
| 66 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.386 |
-0.281 |
| 67 |
253796_at |
AT4G28460
|
unknown |
0.386 |
0.318 |
| 68 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.377 |
-0.249 |
| 69 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
0.374 |
-0.244 |
| 70 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.373 |
-0.908 |
| 71 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.370 |
-0.060 |
| 72 |
249987_at |
AT5G18490
|
unknown |
0.370 |
0.560 |
| 73 |
253992_at |
AT4G26060
|
[Ribosomal protein L18ae family] |
0.369 |
0.019 |
| 74 |
253414_at |
AT4G33050
|
AtIQM1, EDA39, embryo sac development arrest 39, IQM1, IQ-motif protein 1 |
0.363 |
-0.925 |
| 75 |
254487_at |
AT4G20780
|
CML42, calmodulin like 42 |
0.363 |
-0.155 |
| 76 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
0.360 |
-0.855 |
| 77 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.357 |
0.364 |
| 78 |
258787_at |
AT3G11840
|
PUB24, plant U-box 24 |
0.355 |
-0.588 |
| 79 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
0.349 |
0.111 |
| 80 |
258282_at |
AT3G26910
|
[hydroxyproline-rich glycoprotein family protein] |
0.347 |
-0.062 |
| 81 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.347 |
0.355 |
| 82 |
256799_at |
AT3G18560
|
unknown |
0.345 |
0.445 |
| 83 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.339 |
0.252 |
| 84 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
0.335 |
1.109 |
| 85 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
0.333 |
0.771 |
| 86 |
254427_at |
AT4G21190
|
emb1417, embryo defective 1417 |
-0.414 |
0.278 |
| 87 |
261921_at |
AT1G65900
|
unknown |
-0.396 |
1.161 |
| 88 |
256890_at |
AT3G23830
|
AtGRP4, GR-RBP4, GRP4, glycine-rich RNA-binding protein 4 |
-0.379 |
0.448 |
| 89 |
245465_at |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
-0.356 |
-0.095 |
| 90 |
260292_at |
AT1G63680
|
APG13, ALBINO OR PALE-GREEN 13, ATMURE, MURE, PDE316, PIGMENT DEFECTIVE EMBRYO 316 |
-0.343 |
0.629 |
| 91 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
-0.336 |
-0.274 |
| 92 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
-0.333 |
1.553 |
| 93 |
261248_at |
AT1G20030
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.326 |
1.036 |
| 94 |
250528_at |
AT5G08600
|
[U3 ribonucleoprotein (Utp) family protein] |
-0.307 |
0.061 |
| 95 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.304 |
-0.190 |
| 96 |
262236_at |
AT1G48330
|
unknown |
-0.304 |
0.370 |
| 97 |
246746_at |
AT5G27820
|
[Ribosomal L18p/L5e family protein] |
-0.290 |
1.385 |
| 98 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.284 |
-0.916 |
| 99 |
257875_at |
AT3G17120
|
unknown |
-0.280 |
0.668 |
| 100 |
253351_at |
AT4G33700
|
unknown |
-0.273 |
0.111 |
| 101 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
-0.272 |
-0.644 |
| 102 |
248753_at |
AT5G47630
|
mtACP3, mitochondrial acyl carrier protein 3 |
-0.269 |
1.013 |
| 103 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-0.265 |
-1.114 |
| 104 |
251355_at |
AT3G61100
|
[Putative endonuclease or glycosyl hydrolase] |
-0.256 |
0.054 |
| 105 |
266951_at |
AT2G18940
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.250 |
0.020 |
| 106 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.249 |
0.666 |
| 107 |
246797_at |
AT5G26790
|
unknown |
-0.248 |
0.681 |
| 108 |
263688_at |
AT1G26920
|
unknown |
-0.241 |
0.590 |
| 109 |
265730_at |
AT2G32220
|
[Ribosomal L27e protein family] |
-0.238 |
1.005 |
| 110 |
250412_at |
AT5G11150
|
ATVAMP713, vesicle-associated membrane protein 713, VAMP713, VAMP713, vesicle-associated membrane protein 713 |
-0.234 |
0.089 |
| 111 |
262706_at |
AT1G16280
|
AtRH36, Arabidopsis thaliana RNA helicase 36, RH36, RNA helicase 36, SWA3, SLOW WALKER 3 |
-0.232 |
1.227 |
| 112 |
265991_at |
AT2G24120
|
PDE319, PIGMENT DEFECTIVE 319, SCA3, SCABRA 3 |
-0.230 |
0.555 |
| 113 |
266253_at |
AT2G27840
|
HD2D, histone deacetylase 2D, HDA13, HISTONE DEACETYLASE 13, HDT04, HDT4 |
-0.229 |
0.420 |
| 114 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.228 |
0.607 |
| 115 |
263912_at |
AT2G36390
|
BE3, BRANCHING ENZYME 3, SBE2.1, starch branching enzyme 2.1 |
-0.228 |
-0.156 |
| 116 |
248984_at |
AT5G45140
|
NRPC2, nuclear RNA polymerase C2 |
-0.227 |
0.766 |
| 117 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.225 |
0.726 |
| 118 |
253021_at |
AT4G38050
|
[Xanthine/uracil permease family protein] |
-0.224 |
0.868 |
| 119 |
264142_at |
AT1G78930
|
[Mitochondrial transcription termination factor family protein] |
-0.219 |
-0.075 |
| 120 |
260781_at |
AT1G14620
|
DECOY, DECOY |
-0.217 |
0.865 |
| 121 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
-0.213 |
0.840 |
| 122 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
-0.213 |
0.541 |
| 123 |
250744_at |
AT5G05840
|
unknown |
-0.212 |
-0.266 |
| 124 |
258232_at |
AT3G27750
|
EMB3123, EMBRYO DEFECTIVE 3123, THA8, Thylakoid Assembly 8 |
-0.208 |
0.419 |
| 125 |
263548_at |
AT2G21660
|
ATGRP7, GLYCINE RICH PROTEIN 7, CCR2, cold, circadian rhythm, and rna binding 2, GR-RBP7, GLYCINE-RICH RNA-BINDING PROTEIN 7, GRP7, GLYCINE-RICH RNA-BINDING PROTEIN 7 |
-0.207 |
2.065 |
| 126 |
261549_at |
AT1G63470
|
AHL5, AT-hook motif nuclear localized protein 5 |
-0.205 |
0.265 |
| 127 |
254080_at |
AT4G25630
|
ATFIB2, FIB2, fibrillarin 2 |
-0.201 |
0.170 |
| 128 |
261603_at |
AT1G49600
|
ATRBP47A, RNA-binding protein 47A, RBP47A, RNA-binding protein 47A |
-0.201 |
1.518 |
| 129 |
264731_at |
AT1G62150
|
[Mitochondrial transcription termination factor family protein] |
-0.198 |
-0.206 |
| 130 |
260489_at |
AT1G51610
|
[Cation efflux family protein] |
-0.198 |
1.088 |
| 131 |
253092_at |
AT4G37380
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.194 |
-0.363 |
| 132 |
252143_at |
AT3G51150
|
[ATP binding microtubule motor family protein] |
-0.189 |
0.188 |
| 133 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
-0.185 |
-0.017 |
| 134 |
247036_at |
AT5G67130
|
[PLC-like phosphodiesterases superfamily protein] |
-0.184 |
0.172 |
| 135 |
245644_at |
AT1G25320
|
[Leucine-rich repeat protein kinase family protein] |
-0.184 |
0.620 |
| 136 |
259942_at |
AT1G71260
|
ATWHY2, WHIRLY 2, WHY2, WHIRLY 2 |
-0.184 |
1.513 |
| 137 |
263864_at |
AT2G04530
|
CPZ, TRZ2, TRNASE Z 2 |
-0.183 |
1.131 |
| 138 |
251271_at |
AT3G62050
|
[Putative endonuclease or glycosyl hydrolase] |
-0.182 |
0.024 |
| 139 |
258475_at |
AT3G02660
|
EMB2768, EMBRYO DEFECTIVE 2768, FAC31, EMBRYONIC FACTOR 31 |
-0.182 |
0.460 |
| 140 |
247224_at |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
-0.180 |
-0.202 |
| 141 |
267174_at |
AT2G37600
|
[Ribosomal protein L36e family protein] |
-0.180 |
1.024 |
| 142 |
251864_at |
AT3G54920
|
PMR6, powdery mildew resistant 6 |
-0.179 |
0.649 |
| 143 |
245502_at |
AT4G15640
|
unknown |
-0.178 |
-0.138 |
| 144 |
251699_at |
AT3G56630
|
CYP94D2, cytochrome P450, family 94, subfamily D, polypeptide 2 |
-0.178 |
-0.880 |
| 145 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
-0.176 |
0.364 |
| 146 |
260677_at |
AT1G07910
|
AtRLG1, AtRLG1, ATRNL, ARABIDOPSIS THALIANA RNA LIGASE, RNL, RNAligase |
-0.174 |
-0.120 |
| 147 |
263083_at |
AT2G27190
|
ATPAP1, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 1, ATPAP12, PURPLE ACID PHOSPHATASE 12, PAP1, PURPLE ACID PHOSPHATASE 1, PAP12, purple acid phosphatase 12 |
-0.173 |
1.294 |
| 148 |
265383_at |
AT2G16780
|
MSI02, MSI2, MULTICOPY SUPPRESSOR OF IRA1 2, NFC02, NFC2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 2 |
-0.173 |
1.345 |
| 149 |
247461_at |
AT5G62100
|
ATBAG2, BCL-2-associated athanogene 2, BAG2, BCL-2-associated athanogene 2 |
-0.172 |
-1.030 |
| 150 |
250546_at |
AT5G08180
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
-0.172 |
1.353 |
| 151 |
249014_at |
AT5G44710
|
unknown |
-0.171 |
0.242 |
| 152 |
253213_at |
AT4G34910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.170 |
-0.048 |
| 153 |
258995_at |
AT3G01790
|
[Ribosomal protein L13 family protein] |
-0.170 |
0.238 |
| 154 |
252942_at |
AT4G39300
|
unknown |
-0.170 |
0.948 |
| 155 |
267421_at |
AT2G35040
|
[AICARFT/IMPCHase bienzyme family protein] |
-0.168 |
0.593 |
| 156 |
245849_at |
AT5G13520
|
[peptidase M1 family protein] |
-0.167 |
0.372 |
| 157 |
253892_at |
AT4G27620
|
unknown |
-0.167 |
0.575 |
| 158 |
252625_at |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
-0.167 |
0.996 |
| 159 |
251611_at |
AT3G57940
|
unknown |
-0.166 |
-0.037 |
| 160 |
261131_at |
AT1G19835
|
unknown |
-0.166 |
0.469 |
| 161 |
251753_at |
AT3G55760
|
unknown |
-0.166 |
0.106 |
| 162 |
253921_at |
AT4G26900
|
AT-HF, HIS HF, HISN4 |
-0.165 |
-0.009 |
| 163 |
248283_at |
AT5G52920
|
PKP-BETA1, plastidic pyruvate kinase beta subunit 1, PKP1, PLASTIDIAL PYRUVATE KINASE 1, PKP2, PLASTIDIAL PYRUVATE KINASE 2 |
-0.164 |
0.373 |
| 164 |
262818_at |
AT1G11755
|
LEW1, LEAF WILTING 1 |
-0.164 |
-0.087 |
| 165 |
257619_at |
AT3G24810
|
ICK3, KRP5, kip-related protein 5 |
-0.164 |
-0.007 |
| 166 |
246572_at |
AT5G15010
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.160 |
-0.495 |
| 167 |
245106_at |
AT2G41650
|
unknown |
-0.160 |
0.252 |
| 168 |
250326_at |
AT5G12080
|
ATMSL10, MSL10, mechanosensitive channel of small conductance-like 10 |
-0.160 |
0.219 |
| 169 |
259761_at |
AT1G77590
|
LACS9, long chain acyl-CoA synthetase 9 |
-0.160 |
0.684 |