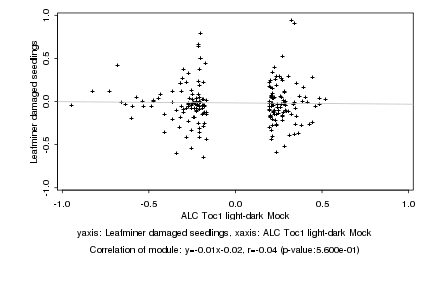
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ALC Toc1 light-dark Mock |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
0.516 |
0.024 |
| 2 |
247518_at |
AT5G61800
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.485 |
-0.034 |
| 3 |
263742_at |
AT2G20625
|
unknown |
0.483 |
0.045 |
| 4 |
262470_at |
AT1G50180
|
[NB-ARC domain-containing disease resistance protein] |
0.459 |
-0.057 |
| 5 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
0.442 |
-0.236 |
| 6 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.440 |
0.285 |
| 7 |
256125_at |
AT1G18250
|
ATLP-1 |
0.422 |
-0.255 |
| 8 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.416 |
-0.003 |
| 9 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.399 |
0.048 |
| 10 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
0.388 |
0.171 |
| 11 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.387 |
0.003 |
| 12 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.379 |
-0.270 |
| 13 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
0.365 |
0.068 |
| 14 |
253270_at |
AT4G34160
|
CYCD3, CYCD3;1, CYCLIN D3;1 |
0.359 |
-0.367 |
| 15 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.349 |
0.212 |
| 16 |
249639_at |
AT5G36930
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.347 |
-0.170 |
| 17 |
260565_at |
AT2G43800
|
[Actin-binding FH2 (formin homology 2) family protein] |
0.345 |
-0.080 |
| 18 |
263485_at |
AT2G29890
|
ATVLN1, VLN1, villin 1 |
0.342 |
-0.266 |
| 19 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.341 |
-0.001 |
| 20 |
255527_at |
AT4G02360
|
unknown |
0.339 |
0.911 |
| 21 |
254544_at |
AT4G19820
|
[Glycosyl hydrolase family protein with chitinase insertion domain] |
0.338 |
-0.375 |
| 22 |
258115_at |
AT3G14670
|
unknown |
0.335 |
-0.030 |
| 23 |
245800_at |
AT1G46264
|
AT-HSFB4, HSFB4, heat shock transcription factor B4, SCZ, SCHIZORIZA |
0.323 |
-0.146 |
| 24 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.318 |
0.951 |
| 25 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
0.309 |
-0.383 |
| 26 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
0.305 |
-0.111 |
| 27 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.303 |
0.292 |
| 28 |
254586_at |
AT4G19510
|
[Disease resistance protein (TIR-NBS-LRR class)] |
0.290 |
-0.113 |
| 29 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
0.288 |
-0.043 |
| 30 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
0.286 |
-0.099 |
| 31 |
250778_at |
AT5G05500
|
MOP10 |
0.285 |
0.001 |
| 32 |
257952_at |
AT3G21770
|
[Peroxidase superfamily protein] |
0.284 |
-0.040 |
| 33 |
253753_at |
AT4G29030
|
[Putative membrane lipoprotein] |
0.282 |
0.122 |
| 34 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.281 |
-0.516 |
| 35 |
254327_at |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.280 |
-0.003 |
| 36 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.278 |
0.108 |
| 37 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
0.278 |
-0.028 |
| 38 |
265993_at |
AT2G24160
|
[pseudogene] |
0.275 |
0.020 |
| 39 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
0.274 |
-0.121 |
| 40 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
0.270 |
0.250 |
| 41 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
0.270 |
-0.160 |
| 42 |
251982_at |
AT3G53190
|
[Pectin lyase-like superfamily protein] |
0.269 |
-0.212 |
| 43 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
0.268 |
0.529 |
| 44 |
249332_at |
AT5G40980
|
unknown |
0.266 |
-0.173 |
| 45 |
251826_at |
AT3G55110
|
ABCG18, ATP-binding cassette G18 |
0.265 |
0.273 |
| 46 |
255546_at |
AT4G01910
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.263 |
0.050 |
| 47 |
248079_at |
AT5G55790
|
unknown |
0.262 |
-0.024 |
| 48 |
258118_at |
AT3G14710
|
[RNI-like superfamily protein] |
0.262 |
-0.024 |
| 49 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
0.257 |
-0.058 |
| 50 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
0.255 |
0.068 |
| 51 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.253 |
0.291 |
| 52 |
248977_at |
AT5G45020
|
[Glutathione S-transferase family protein] |
0.248 |
-0.102 |
| 53 |
261460_at |
AT1G07880
|
ATMPK13 |
0.248 |
-0.131 |
| 54 |
246906_at |
AT5G25475
|
[AP2/B3-like transcriptional factor family protein] |
0.240 |
-0.032 |
| 55 |
266735_at |
AT2G46930
|
[Pectinacetylesterase family protein] |
0.239 |
-0.065 |
| 56 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
0.237 |
-0.267 |
| 57 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.237 |
-0.142 |
| 58 |
258893_at |
AT3G05660
|
AtRLP33, receptor like protein 33, RLP33, receptor like protein 33 |
0.235 |
0.195 |
| 59 |
246584_at |
AT5G14730
|
unknown |
0.233 |
0.298 |
| 60 |
248427_at |
AT5G51750
|
ATSBT1.3, subtilase 1.3, SBT1.3, subtilase 1.3 |
0.233 |
-0.265 |
| 61 |
252916_at |
AT4G38950
|
[ATP binding microtubule motor family protein] |
0.232 |
-0.588 |
| 62 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.231 |
0.259 |
| 63 |
259166_at |
AT3G01670
|
AtSEOR2, Arabidopsis thaliana sieve element occlusion-related 2, SEOa, sieve element occlusion a, SEOR2, sieve element occlusion-related 2 |
0.226 |
-0.220 |
| 64 |
250565_at |
AT5G08000
|
E13L3, glucan endo-1,3-beta-glucosidase-like protein 3, PDCB2, PLASMODESMATA CALLOSE-BINDING PROTEIN 2 |
0.224 |
-0.149 |
| 65 |
260991_at |
AT1G12160
|
[Flavin-binding monooxygenase family protein] |
0.223 |
0.056 |
| 66 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.220 |
0.402 |
| 67 |
256454_at |
AT1G75280
|
[NmrA-like negative transcriptional regulator family protein] |
0.220 |
-0.095 |
| 68 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
0.216 |
-0.043 |
| 69 |
257191_at |
AT3G13175
|
unknown |
0.216 |
0.043 |
| 70 |
250109_at |
AT5G15230
|
GASA4, GAST1 protein homolog 4 |
0.215 |
-0.109 |
| 71 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
0.215 |
-0.127 |
| 72 |
249214_at |
AT5G42720
|
[Glycosyl hydrolase family 17 protein] |
0.214 |
-0.396 |
| 73 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
0.214 |
0.041 |
| 74 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.214 |
0.097 |
| 75 |
264200_at |
AT1G22650
|
A/N-InvD, alkaline/neutral invertase D |
0.214 |
-0.014 |
| 76 |
255236_at |
AT4G05520
|
ATEHD2, EPS15 homology domain 2, EHD2, EPS15 homology domain 2 |
0.213 |
-0.014 |
| 77 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
0.212 |
-0.273 |
| 78 |
266223_at |
AT2G28790
|
[Pathogenesis-related thaumatin superfamily protein] |
0.212 |
0.342 |
| 79 |
262753_at |
AT1G16340
|
ATKDSA2, ATKSDA |
0.209 |
-0.202 |
| 80 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
0.209 |
0.159 |
| 81 |
262206_at |
AT2G01090
|
[Ubiquinol-cytochrome C reductase hinge protein] |
0.208 |
-0.020 |
| 82 |
249115_at |
AT5G43810
|
AGO10, ARGONAUTE 10, PNH, PINHEAD, ZLL, ZWILLE |
0.207 |
-0.336 |
| 83 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.206 |
0.048 |
| 84 |
263913_at |
AT2G36570
|
PXC1, PXY/TDR-correlated 1 |
0.206 |
-0.432 |
| 85 |
266838_at |
AT2G25980
|
[Mannose-binding lectin superfamily protein] |
0.206 |
0.075 |
| 86 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
0.204 |
0.060 |
| 87 |
253740_at |
AT4G28706
|
[pfkB-like carbohydrate kinase family protein] |
0.204 |
-0.047 |
| 88 |
260144_at |
AT1G71960
|
ABCG25, ATP-binding casette G25, ATABCG25, Arabidopsis thaliana ATP-binding cassette G25 |
0.203 |
0.053 |
| 89 |
255484_at |
AT4G02540
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.201 |
-0.155 |
| 90 |
266104_at |
AT2G45150
|
CDS4, cytidinediphosphate diacylglycerol synthase 4 |
0.199 |
-0.170 |
| 91 |
247948_at |
AT5G57130
|
SMXL5, SMAX1-like 5 |
0.198 |
-0.181 |
| 92 |
260012_at |
AT1G67865
|
unknown |
0.198 |
0.169 |
| 93 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.198 |
0.245 |
| 94 |
260112_at |
AT1G63310
|
unknown |
0.198 |
-0.168 |
| 95 |
267627_at |
AT2G42270
|
[U5 small nuclear ribonucleoprotein helicase] |
0.196 |
-0.099 |
| 96 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.195 |
-0.050 |
| 97 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
0.193 |
0.180 |
| 98 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.193 |
0.008 |
| 99 |
254532_at |
AT4G19660
|
ATNPR4, NPR4, NPR1-like protein 4 |
0.193 |
-0.296 |
| 100 |
261879_at |
AT1G50520
|
CYP705A27, cytochrome P450, family 705, subfamily A, polypeptide 27 |
0.193 |
-0.022 |
| 101 |
263465_at |
AT2G31940
|
unknown |
0.191 |
0.225 |
| 102 |
260181_at |
AT1G70710
|
ATGH9B1, glycosyl hydrolase 9B1, CEL1, CELLULASE 1, GH9B1, glycosyl hydrolase 9B1 |
0.191 |
-0.081 |
| 103 |
248810_at |
AT5G47280
|
ADR1-L3, ADR1-like 3 |
-0.949 |
-0.036 |
| 104 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.830 |
0.123 |
| 105 |
260396_at |
AT1G69720
|
HO3, heme oxygenase 3 |
-0.732 |
0.125 |
| 106 |
252345_at |
AT3G48640
|
unknown |
-0.682 |
0.420 |
| 107 |
250135_at |
AT5G15360
|
unknown |
-0.661 |
-0.011 |
| 108 |
255414_at |
AT4G03156
|
[small GTPase-related] |
-0.638 |
-0.028 |
| 109 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.601 |
-0.190 |
| 110 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
-0.599 |
-0.053 |
| 111 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
-0.574 |
0.055 |
| 112 |
254566_at |
AT4G19240
|
unknown |
-0.541 |
0.009 |
| 113 |
257417_at |
AT1G10110
|
[F-box family protein] |
-0.533 |
-0.054 |
| 114 |
253689_at |
AT4G29770
|
unknown |
-0.487 |
-0.047 |
| 115 |
252663_at |
AT3G44070
|
[Glycosyl hydrolase family 35 protein] |
-0.474 |
0.002 |
| 116 |
256368_at |
AT1G66800
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.474 |
0.021 |
| 117 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.447 |
0.039 |
| 118 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
-0.433 |
0.089 |
| 119 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.413 |
-0.142 |
| 120 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.412 |
-0.348 |
| 121 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
-0.369 |
0.120 |
| 122 |
262421_at |
AT1G50290
|
unknown |
-0.367 |
-0.009 |
| 123 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.364 |
-0.208 |
| 124 |
254350_at |
AT4G22280
|
[F-box/RNI-like superfamily protein] |
-0.345 |
-0.104 |
| 125 |
250936_at |
AT5G03120
|
unknown |
-0.341 |
-0.598 |
| 126 |
251058_at |
AT5G01790
|
unknown |
-0.327 |
-0.294 |
| 127 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
-0.319 |
0.210 |
| 128 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
-0.319 |
0.212 |
| 129 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
-0.318 |
-0.180 |
| 130 |
265327_at |
AT2G18210
|
unknown |
-0.315 |
0.117 |
| 131 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.313 |
-0.049 |
| 132 |
246888_at |
AT5G26270
|
unknown |
-0.308 |
0.273 |
| 133 |
247250_at |
AT5G64630
|
FAS2, FASCIATA 2, MUB3.9, NFB01, NFB1 |
-0.305 |
-0.082 |
| 134 |
250772_at |
AT5G05420
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.303 |
-0.125 |
| 135 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.301 |
0.381 |
| 136 |
254683_at |
AT4G13800
|
unknown |
-0.285 |
0.221 |
| 137 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.285 |
-0.415 |
| 138 |
245227_s_at |
AT1G08410
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.283 |
-0.071 |
| 139 |
257817_at |
AT3G25150
|
[Nuclear transport factor 2 (NTF2) family protein with RNA binding (RRM-RBD-RNP motifs) domain] |
-0.276 |
-0.031 |
| 140 |
252204_at |
AT3G50340
|
unknown |
-0.275 |
-0.013 |
| 141 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.274 |
-0.227 |
| 142 |
264773_at |
AT1G22900
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.272 |
0.335 |
| 143 |
246776_at |
AT5G27550
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.272 |
-0.056 |
| 144 |
259661_at |
AT1G55265
|
unknown |
-0.271 |
0.044 |
| 145 |
253052_at |
AT4G37310
|
CYP81H1, cytochrome P450, family 81, subfamily H, polypeptide 1 |
-0.258 |
-0.044 |
| 146 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
-0.258 |
-0.333 |
| 147 |
259528_at |
AT1G12330
|
unknown |
-0.258 |
-0.534 |
| 148 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
-0.258 |
-0.079 |
| 149 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.255 |
0.128 |
| 150 |
250773_at |
AT5G05430
|
[RNA-binding protein] |
-0.251 |
0.031 |
| 151 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
-0.251 |
-0.012 |
| 152 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
-0.250 |
0.091 |
| 153 |
245240_at |
AT1G44510
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.246 |
-0.003 |
| 154 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.245 |
-0.177 |
| 155 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.240 |
-0.182 |
| 156 |
256976_at |
AT3G21020
|
[copia-like retrotransposon family, has a 5.4e-14 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-0.240 |
-0.079 |
| 157 |
254596_at |
AT4G18975
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.238 |
-0.016 |
| 158 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.232 |
-0.032 |
| 159 |
256832_at |
AT3G22880
|
ARLIM15, ARABIDOPSIS HOMOLOG OF LILY MESSAGES INDUCED AT MEIOSIS 15, ATDMC1, ARABIDOPSIS THALIANA DISRUPTION OF MEIOTIC CONTROL 1, DMC1, DISRUPTION OF MEIOTIC CONTROL 1 |
-0.230 |
-0.107 |
| 160 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
-0.229 |
-0.096 |
| 161 |
254573_at |
AT4G19420
|
[Pectinacetylesterase family protein] |
-0.228 |
0.046 |
| 162 |
249127_at |
AT5G43500
|
ARP9, actin-related protein 9, ATARP9, actin-related protein 9 |
-0.227 |
-0.070 |
| 163 |
246326_at |
AT1G16590
|
ATREV7, REV7 |
-0.225 |
0.003 |
| 164 |
247800_at |
AT5G58570
|
unknown |
-0.224 |
-0.044 |
| 165 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.221 |
-0.031 |
| 166 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.217 |
0.639 |
| 167 |
254292_at |
AT4G23030
|
[MATE efflux family protein] |
-0.217 |
0.234 |
| 168 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
-0.217 |
0.669 |
| 169 |
253412_at |
AT4G33000
|
ATCBL10, CBL10, calcineurin B-like protein 10, SCABP8, SOS3-LIKE CALCIUM BINDING PROTEIN 8 |
-0.216 |
-0.245 |
| 170 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
-0.215 |
0.090 |
| 171 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.213 |
-0.416 |
| 172 |
263741_at |
AT2G20620
|
unknown |
-0.212 |
-0.087 |
| 173 |
262870_at |
AT1G64710
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.211 |
0.190 |
| 174 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.210 |
-0.305 |
| 175 |
266355_at |
AT2G01400
|
unknown |
-0.210 |
0.047 |
| 176 |
264252_at |
AT1G09180
|
ATSAR1, SECRETION-ASSOCIATED RAS 1, ATSARA1A, secretion-associated RAS super family 1, SARA1A, secretion-associated RAS super family 1 |
-0.208 |
0.380 |
| 177 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.208 |
-0.348 |
| 178 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-0.208 |
-0.047 |
| 179 |
258802_at |
AT3G04650
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.208 |
0.011 |
| 180 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.208 |
-0.005 |
| 181 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
-0.206 |
0.795 |
| 182 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
-0.204 |
0.500 |
| 183 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
-0.204 |
0.509 |
| 184 |
250882_at |
AT5G04000
|
unknown |
-0.202 |
0.000 |
| 185 |
266824_at |
AT2G22800
|
HAT9 |
-0.195 |
-0.042 |
| 186 |
259683_at |
AT1G63050
|
AtLPLAT2, LPCAT2, lysophosphatidylcholine acyltransferase 2, LPLAT2, lysophospholipid acyltransferase 2 |
-0.192 |
-0.072 |
| 187 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.191 |
-0.143 |
| 188 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
-0.191 |
0.030 |
| 189 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.190 |
0.029 |
| 190 |
265428_at |
AT2G20720
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.188 |
-0.037 |
| 191 |
254145_at |
AT4G24700
|
unknown |
-0.188 |
-0.645 |
| 192 |
254524_at |
AT4G20000
|
[VQ motif-containing protein] |
-0.187 |
0.228 |
| 193 |
262780_at |
AT1G13090
|
CYP71B28, cytochrome P450, family 71, subfamily B, polypeptide 28 |
-0.186 |
-0.135 |
| 194 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
-0.185 |
-0.288 |
| 195 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
-0.184 |
-0.119 |
| 196 |
258581_at |
AT3G04160
|
unknown |
-0.182 |
-0.056 |
| 197 |
258283_at |
AT3G26750
|
unknown |
-0.180 |
-0.036 |
| 198 |
266402_at |
AT2G38780
|
unknown |
-0.179 |
-0.246 |
| 199 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
-0.173 |
0.445 |
| 200 |
257610_at |
AT3G13810
|
AtIDD11, indeterminate(ID)-domain 11, IDD11, indeterminate(ID)-domain 11 |
-0.172 |
-0.123 |
| 201 |
259111_at |
AT3G05520
|
AtCPA, CPA, capping protein A |
-0.171 |
0.015 |
| 202 |
254221_at |
AT4G23820
|
[Pectin lyase-like superfamily protein] |
-0.171 |
-0.431 |
| 203 |
253945_at |
AT4G27050
|
[F-box/RNI-like superfamily protein] |
-0.169 |
-0.140 |