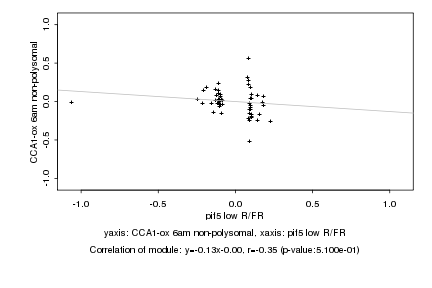
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
pif5 low R/FR |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.221 |
-0.255 |
| 2 |
260012_at |
AT1G67865
|
unknown |
0.179 |
-0.049 |
| 3 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
0.177 |
0.077 |
| 4 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.172 |
-0.001 |
| 5 |
263260_at |
AT1G10570
|
OTS2, OVERLY TOLERANT TO SALT 2, ULP1C, UB-LIKE PROTEASE 1C |
0.155 |
-0.156 |
| 6 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
0.140 |
-0.236 |
| 7 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
0.139 |
0.085 |
| 8 |
247366_at |
AT5G63260
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.104 |
-0.161 |
| 9 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
0.103 |
-0.193 |
| 10 |
264534_at |
AT1G55700
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.103 |
0.103 |
| 11 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
0.101 |
-0.200 |
| 12 |
256096_at |
AT1G13650
|
unknown |
0.100 |
0.043 |
| 13 |
257381_at |
AT2G37950
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.094 |
-0.101 |
| 14 |
251294_at |
AT3G61940
|
ATMTPA1, MTPA1 |
0.093 |
-0.072 |
| 15 |
264241_at |
AT1G54840
|
[HSP20-like chaperones superfamily protein] |
0.093 |
-0.040 |
| 16 |
258380_at |
AT3G16650
|
PRL2, Pleiotropic regulatory locus 2 |
0.092 |
0.185 |
| 17 |
245128_at |
AT2G45380
|
unknown |
0.092 |
0.043 |
| 18 |
257668_at |
AT3G20460
|
[Major facilitator superfamily protein] |
0.086 |
-0.243 |
| 19 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
0.085 |
-0.100 |
| 20 |
255750_at |
AT1G31960
|
unknown |
0.085 |
-0.146 |
| 21 |
260744_at |
AT1G15010
|
unknown |
0.085 |
-0.016 |
| 22 |
260004_at |
AT1G67860
|
unknown |
0.085 |
-0.520 |
| 23 |
259928_at |
AT1G34380
|
[5'-3' exonuclease family protein] |
0.083 |
0.285 |
| 24 |
255346_at |
AT4G03800
|
[gypsy-like retrotransposon family, has a 1.5e-125 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
0.083 |
0.229 |
| 25 |
263329_at |
AT2G15260
|
[RING/U-box superfamily protein] |
0.080 |
-0.217 |
| 26 |
261501_at |
AT1G28390
|
[Protein kinase superfamily protein] |
0.079 |
0.563 |
| 27 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.078 |
0.316 |
| 28 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
-1.067 |
-0.012 |
| 29 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
-0.249 |
0.035 |
| 30 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
-0.214 |
-0.025 |
| 31 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
-0.208 |
0.152 |
| 32 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.192 |
0.192 |
| 33 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
-0.156 |
-0.019 |
| 34 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.144 |
-0.141 |
| 35 |
257502_at |
AT1G78110
|
unknown |
-0.134 |
0.167 |
| 36 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.133 |
0.014 |
| 37 |
262244_at |
AT1G48260
|
CIPK17, CBL-interacting protein kinase 17, SnRK3.21, SNF1-RELATED PROTEIN KINASE 3.21 |
-0.125 |
0.081 |
| 38 |
260819_at |
AT1G06850
|
AtbZIP52, basic leucine-zipper 52, bZIP52, basic leucine-zipper 52 |
-0.117 |
-0.023 |
| 39 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.114 |
0.111 |
| 40 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
-0.113 |
0.031 |
| 41 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.112 |
0.237 |
| 42 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.111 |
-0.015 |
| 43 |
245384_at |
AT4G16790
|
[hydroxyproline-rich glycoprotein family protein] |
-0.110 |
0.146 |
| 44 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.109 |
-0.058 |
| 45 |
247690_at |
AT5G59800
|
ATMBD7, ARABIDOPSIS THALIANA METHYL-CPG-BINDING DOMAIN 7, MBD7, methyl-CPG-binding domain 7 |
-0.109 |
0.007 |
| 46 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.105 |
0.030 |
| 47 |
246888_at |
AT5G26270
|
unknown |
-0.102 |
0.099 |
| 48 |
266654_at |
AT2G25890
|
[Oleosin family protein] |
-0.101 |
0.073 |
| 49 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.100 |
-0.032 |
| 50 |
254566_at |
AT4G19240
|
unknown |
-0.095 |
-0.149 |
| 51 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.095 |
0.039 |
| 52 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.090 |
-0.034 |
| 53 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
-0.090 |
0.018 |