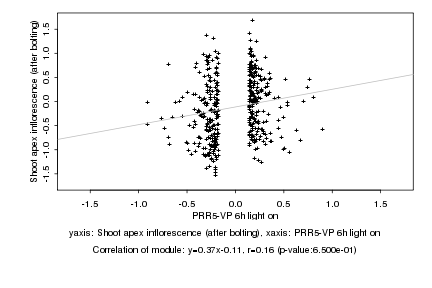
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
PRR5-VP 6h light on |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
0.893 |
-0.573 |
| 2 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.799 |
0.087 |
| 3 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.763 |
0.464 |
| 4 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.736 |
0.303 |
| 5 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.693 |
0.006 |
| 6 |
251793_at |
AT3G55580
|
[Regulator of chromosome condensation (RCC1) family protein] |
0.662 |
-0.788 |
| 7 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.628 |
-0.599 |
| 8 |
258893_at |
AT3G05660
|
AtRLP33, receptor like protein 33, RLP33, receptor like protein 33 |
0.552 |
-1.041 |
| 9 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.530 |
-0.063 |
| 10 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
0.530 |
-0.002 |
| 11 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.516 |
0.458 |
| 12 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
0.506 |
-0.964 |
| 13 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.493 |
-0.992 |
| 14 |
256114_at |
AT1G16850
|
unknown |
0.491 |
-0.312 |
| 15 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.474 |
-0.727 |
| 16 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.459 |
-0.111 |
| 17 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.442 |
-0.379 |
| 18 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
0.441 |
-0.552 |
| 19 |
257057_at |
AT3G15310
|
unknown |
0.438 |
0.093 |
| 20 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.399 |
0.067 |
| 21 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
0.371 |
-0.810 |
| 22 |
251062_at |
AT5G01840
|
ATOFP1, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 1, OFP1, ovate family protein 1 |
0.358 |
-0.816 |
| 23 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
0.358 |
-0.653 |
| 24 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.355 |
0.494 |
| 25 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.348 |
0.195 |
| 26 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.346 |
0.582 |
| 27 |
261318_at |
AT1G53035
|
unknown |
0.342 |
0.475 |
| 28 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
0.339 |
0.163 |
| 29 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
0.333 |
-0.267 |
| 30 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.322 |
0.374 |
| 31 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.321 |
0.324 |
| 32 |
264604_at |
AT1G04650
|
unknown |
0.317 |
0.139 |
| 33 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
0.314 |
-0.409 |
| 34 |
258008_at |
AT3G19430
|
[late embryogenesis abundant protein-related / LEA protein-related] |
0.311 |
-0.755 |
| 35 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.305 |
0.916 |
| 36 |
264597_at |
AT1G04620
|
HCAR, 7-hydroxymethyl chlorophyll a (HMChl) reductase |
0.305 |
0.294 |
| 37 |
260688_at |
AT1G17665
|
unknown |
0.304 |
-0.876 |
| 38 |
246537_at |
AT5G15940
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.304 |
-0.684 |
| 39 |
250194_at |
AT5G14550
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.304 |
0.180 |
| 40 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
0.297 |
0.480 |
| 41 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
0.297 |
0.472 |
| 42 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
0.288 |
-0.813 |
| 43 |
252916_at |
AT4G38950
|
[ATP binding microtubule motor family protein] |
0.287 |
-0.420 |
| 44 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.286 |
-0.201 |
| 45 |
256131_at |
AT1G13600
|
AtbZIP58, basic leucine-zipper 58, bZIP58, basic leucine-zipper 58 |
0.282 |
-0.656 |
| 46 |
261754_at |
AT1G76130
|
AMY2, alpha-amylase-like 2, ATAMY2, ARABIDOPSIS THALIANA ALPHA-AMYLASE-LIKE 2 |
0.281 |
-0.607 |
| 47 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
0.279 |
-0.577 |
| 48 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.269 |
0.266 |
| 49 |
254085_at |
AT4G24960
|
ATHVA22D, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE D, HVA22D, HVA22 homologue D |
0.266 |
0.679 |
| 50 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
0.265 |
0.437 |
| 51 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
0.265 |
-0.562 |
| 52 |
261545_at |
AT1G63530
|
unknown |
0.262 |
-1.260 |
| 53 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
0.259 |
0.211 |
| 54 |
261187_at |
AT1G32860
|
[Glycosyl hydrolase superfamily protein] |
0.259 |
0.230 |
| 55 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
0.258 |
0.529 |
| 56 |
262124_at |
AT1G59660
|
[Nucleoporin autopeptidase] |
0.255 |
0.472 |
| 57 |
250412_at |
AT5G11150
|
ATVAMP713, vesicle-associated membrane protein 713, VAMP713, VAMP713, vesicle-associated membrane protein 713 |
0.250 |
0.074 |
| 58 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.250 |
-0.725 |
| 59 |
260264_at |
AT1G68500
|
unknown |
0.240 |
-0.773 |
| 60 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.235 |
0.694 |
| 61 |
266663_at |
AT2G25790
|
SKM1, STERILITY-REGULATING KINASE MEMBER 1 |
0.235 |
-0.444 |
| 62 |
254174_at |
AT4G24120
|
ATYSL1, YELLOW STRIPE LIKE 1, YSL1, YELLOW STRIPE like 1 |
0.234 |
0.555 |
| 63 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
0.233 |
0.099 |
| 64 |
253679_at |
AT4G29610
|
[Cytidine/deoxycytidylate deaminase family protein] |
0.232 |
-0.558 |
| 65 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.231 |
0.189 |
| 66 |
251896_at |
AT3G54390
|
[sequence-specific DNA binding transcription factors] |
0.231 |
-0.394 |
| 67 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.230 |
-0.199 |
| 68 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
0.228 |
-1.205 |
| 69 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
0.227 |
0.017 |
| 70 |
251753_at |
AT3G55760
|
unknown |
0.225 |
0.291 |
| 71 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
0.220 |
-0.962 |
| 72 |
264635_at |
AT1G65500
|
unknown |
0.220 |
0.393 |
| 73 |
245090_at |
AT2G40900
|
UMAMIT11, Usually multiple acids move in and out Transporters 11 |
0.220 |
-0.432 |
| 74 |
267307_at |
AT2G30210
|
LAC3, laccase 3 |
0.219 |
-0.603 |
| 75 |
251870_at |
AT3G54510
|
[Early-responsive to dehydration stress protein (ERD4)] |
0.219 |
0.169 |
| 76 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.217 |
-0.771 |
| 77 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
0.216 |
0.245 |
| 78 |
254563_at |
AT4G19120
|
ERD3, early-responsive to dehydration 3 |
0.215 |
0.695 |
| 79 |
247937_at |
AT5G57110
|
ACA8, autoinhibited Ca2+ -ATPase, isoform 8, AT-ACA8, AUTOINHIBITED CA2+ -ATPASE, ISOFORM 8 |
0.214 |
0.051 |
| 80 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
0.214 |
0.544 |
| 81 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.213 |
0.870 |
| 82 |
261048_at |
AT1G01420
|
UGT72B3, UDP-glucosyl transferase 72B3 |
0.212 |
0.016 |
| 83 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
0.209 |
-0.209 |
| 84 |
264186_at |
AT1G54570
|
PES1, phytyl ester synthase 1 |
0.209 |
-0.271 |
| 85 |
254515_at |
AT4G20270
|
BAM3, BARELY ANY MERISTEM 3 |
0.209 |
1.254 |
| 86 |
247645_at |
AT5G60530
|
[late embryogenesis abundant protein-related / LEA protein-related] |
0.208 |
-0.676 |
| 87 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
0.208 |
-0.029 |
| 88 |
261366_at |
AT1G53100
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.205 |
-0.412 |
| 89 |
264091_at |
AT1G79110
|
BRG2, BOI-related gene 2 |
0.205 |
0.611 |
| 90 |
257237_at |
AT3G14890
|
[phosphoesterase] |
0.202 |
0.757 |
| 91 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.201 |
-0.986 |
| 92 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.200 |
-0.827 |
| 93 |
253253_at |
AT4G34750
|
SAUR49, SMALL AUXIN UPREGULATED RNA 49 |
0.200 |
0.749 |
| 94 |
259303_at |
AT3G05130
|
unknown |
0.199 |
-0.555 |
| 95 |
251190_at |
AT3G62690
|
ATL5, AtL5 |
0.199 |
0.065 |
| 96 |
247147_at |
AT5G65630
|
GTE7, global transcription factor group E7 |
0.197 |
0.629 |
| 97 |
252047_at |
AT3G52490
|
SMXL3, SMAX1-like 3 |
0.197 |
0.030 |
| 98 |
261073_at |
AT1G07300
|
[josephin protein-related] |
0.196 |
-0.847 |
| 99 |
251735_at |
AT3G56090
|
ATFER3, ferritin 3, FER3, ferritin 3 |
0.196 |
0.433 |
| 100 |
264989_at |
AT1G27200
|
unknown |
0.196 |
0.378 |
| 101 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
0.193 |
-0.012 |
| 102 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
0.193 |
0.751 |
| 103 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
0.191 |
-0.773 |
| 104 |
262760_at |
AT1G10770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.191 |
-1.164 |
| 105 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
0.191 |
-0.346 |
| 106 |
259871_at |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
0.190 |
0.075 |
| 107 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.190 |
0.706 |
| 108 |
253056_at |
AT4G37650
|
SGR7, SHOOT GRAVITROPISM 7, SHR, SHORT ROOT |
0.188 |
0.117 |
| 109 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
0.187 |
-0.364 |
| 110 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
0.187 |
0.598 |
| 111 |
251130_at |
AT5G01180
|
AtNPF8.2, ATPTR5, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 5, NPF8.2, NRT1/ PTR family 8.2, PTR5, peptide transporter 5 |
0.186 |
0.353 |
| 112 |
265842_at |
AT2G35700
|
ATERF38, ERF FAMILY PROTEIN 38, ERF38, ERF family protein 38 |
0.186 |
0.135 |
| 113 |
260618_at |
AT1G53230
|
TCP3, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 3 |
0.185 |
0.301 |
| 114 |
246989_at |
AT5G67350
|
unknown |
0.183 |
-0.447 |
| 115 |
250413_at |
AT5G11160
|
APT5, adenine phosphoribosyltransferase 5 |
0.181 |
-0.017 |
| 116 |
251907_at |
AT3G53760
|
ATGCP4, GCP4, GAMMA-TUBULIN COMPLEX PROTEIN 4 |
0.180 |
0.054 |
| 117 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.178 |
0.517 |
| 118 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
0.178 |
0.533 |
| 119 |
253021_at |
AT4G38050
|
[Xanthine/uracil permease family protein] |
0.178 |
0.973 |
| 120 |
263751_at |
AT2G21300
|
[ATP binding microtubule motor family protein] |
0.178 |
0.943 |
| 121 |
265049_at |
AT1G52060
|
[Mannose-binding lectin superfamily protein] |
0.177 |
0.024 |
| 122 |
247776_at |
AT5G58700
|
ATPLC4, PLC4, phosphatidylinositol-speciwc phospholipase C4 |
0.177 |
0.174 |
| 123 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.177 |
-0.705 |
| 124 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
0.176 |
0.552 |
| 125 |
262881_at |
AT1G64890
|
[Major facilitator superfamily protein] |
0.176 |
-0.516 |
| 126 |
266225_at |
AT2G28900
|
ATOEP16-1, outer plastid envelope protein 16-1, ATOEP16-L, OUTER PLASTID ENVELOPE PROTEIN 16-L, OEP16, outer envelope protein 16, OEP16-1, outer plastid envelope protein 16-1 |
0.175 |
1.701 |
| 127 |
245341_at |
AT4G16447
|
unknown |
0.175 |
0.538 |
| 128 |
254575_at |
AT4G19460
|
[UDP-Glycosyltransferase superfamily protein] |
0.174 |
-0.808 |
| 129 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
0.173 |
-0.551 |
| 130 |
259864_at |
AT1G72800
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.173 |
-0.017 |
| 131 |
245699_at |
AT5G04250
|
[Cysteine proteinases superfamily protein] |
0.172 |
-0.238 |
| 132 |
252866_at |
AT4G39840
|
unknown |
0.171 |
0.673 |
| 133 |
264000_at |
AT2G22500
|
ATPUMP5, PLANT UNCOUPLING MITOCHONDRIAL PROTEIN 5, DIC1, DICARBOXYLATE CARRIER 1, UCP5, uncoupling protein 5 |
0.171 |
-0.051 |
| 134 |
250517_at |
AT5G08260
|
scpl35, serine carboxypeptidase-like 35 |
0.170 |
0.160 |
| 135 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.170 |
-0.702 |
| 136 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.169 |
-0.476 |
| 137 |
252917_at |
AT4G38960
|
BBX19, B-box domain protein 19 |
0.167 |
0.699 |
| 138 |
251530_at |
AT3G58520
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
0.167 |
-0.098 |
| 139 |
259411_at |
AT1G13410
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.167 |
-0.249 |
| 140 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
0.167 |
-0.599 |
| 141 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
0.167 |
-0.202 |
| 142 |
267280_at |
AT2G19450
|
ABX45, AS11, ATDGAT, Arabidopsis thaliana acyl-CoA:diacylglycerol acyltransferase, AtDGAT1, DGAT1, acyl-CoA:diacylglycerol acyltransferase 1, RDS1, TAG1, TRIACYLGLYCEROL BIOSYNTHESIS DEFECT 1 |
0.166 |
-0.330 |
| 143 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
0.164 |
0.292 |
| 144 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.164 |
0.753 |
| 145 |
261125_at |
AT1G04990
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.162 |
0.168 |
| 146 |
257724_at |
AT3G18510
|
unknown |
0.161 |
0.814 |
| 147 |
260489_at |
AT1G51610
|
[Cation efflux family protein] |
0.160 |
1.044 |
| 148 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.160 |
-0.698 |
| 149 |
248148_at |
AT5G54930
|
[AT hook motif-containing protein] |
0.160 |
0.617 |
| 150 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.159 |
-0.742 |
| 151 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
0.159 |
-0.745 |
| 152 |
248784_at |
AT5G47380
|
unknown |
0.159 |
0.935 |
| 153 |
250689_at |
AT5G06610
|
unknown |
0.158 |
0.227 |
| 154 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
0.158 |
-0.780 |
| 155 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.158 |
0.195 |
| 156 |
254850_at |
AT4G12000
|
[SNARE associated Golgi protein family] |
0.158 |
0.544 |
| 157 |
265032_at |
AT1G61580
|
ARP2, ARABIDOPSIS RIBOSOMAL PROTEIN 2, RPL3B, R-protein L3 B |
0.158 |
0.731 |
| 158 |
259003_at |
AT3G02010
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.157 |
0.402 |
| 159 |
251075_at |
AT5G01890
|
PXC2, PXY/TDR-correlated 2 |
0.157 |
0.033 |
| 160 |
259394_at |
AT1G06420
|
unknown |
0.157 |
-0.298 |
| 161 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.157 |
0.837 |
| 162 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
0.155 |
0.221 |
| 163 |
246994_at |
AT5G67460
|
[O-Glycosyl hydrolases family 17 protein] |
0.155 |
0.781 |
| 164 |
260150_at |
AT1G52820
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.154 |
-0.608 |
| 165 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.154 |
0.707 |
| 166 |
245533_at |
AT4G15130
|
ATCCT2, CCT2, phosphorylcholine cytidylyltransferase2 |
0.153 |
-0.475 |
| 167 |
249422_at |
AT5G39760
|
AtHB23, homeobox protein 23, HB23, homeobox protein 23, ZHD10, ZINC FINGER HOMEODOMAIN 10 |
0.153 |
0.393 |
| 168 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
0.153 |
0.678 |
| 169 |
266007_at |
AT2G37380
|
MAKR3, MEMBRANE-ASSOCIATED KINASE REGULATOR 3 |
0.153 |
0.057 |
| 170 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
0.152 |
-0.214 |
| 171 |
252359_at |
AT3G48440
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.152 |
1.116 |
| 172 |
261248_at |
AT1G20030
|
[Pathogenesis-related thaumatin superfamily protein] |
0.152 |
1.084 |
| 173 |
257456_at |
AT2G18120
|
SRS4, SHI-related sequence 4 |
0.152 |
0.113 |
| 174 |
249990_at |
AT5G18540
|
unknown |
0.152 |
-0.209 |
| 175 |
253054_at |
AT4G37580
|
COP3, CONSTITUTIVE PHOTOMORPHOGENIC 3, HLS1, HOOKLESS 1, UNS2, UNUSUAL SUGAR RESPONSE 2 |
0.151 |
0.438 |
| 176 |
249612_at |
AT5G37290
|
[ARM repeat superfamily protein] |
0.151 |
0.986 |
| 177 |
246983_at |
AT5G67200
|
[Leucine-rich repeat protein kinase family protein] |
0.150 |
0.129 |
| 178 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.149 |
0.563 |
| 179 |
250053_at |
AT5G17850
|
[Sodium/calcium exchanger family protein] |
0.149 |
-0.336 |
| 180 |
252707_at |
AT3G43790
|
ZIFL2, zinc induced facilitator-like 2 |
0.148 |
0.115 |
| 181 |
265048_at |
AT1G52050
|
[Mannose-binding lectin superfamily protein] |
0.148 |
-0.486 |
| 182 |
246995_at |
AT5G67470
|
ATFH6, ARABIDOPSIS FORMIN HOMOLOG 6, FH6, formin homolog 6 |
0.147 |
0.234 |
| 183 |
266166_at |
AT2G28080
|
[UDP-Glycosyltransferase superfamily protein] |
0.147 |
1.277 |
| 184 |
259856_at |
AT1G68440
|
unknown |
0.145 |
0.381 |
| 185 |
252951_at |
AT4G38700
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.145 |
-0.157 |
| 186 |
261586_at |
AT1G01640
|
[BTB/POZ domain-containing protein] |
0.145 |
-0.099 |
| 187 |
265050_at |
AT1G52070
|
[Mannose-binding lectin superfamily protein] |
0.145 |
-0.688 |
| 188 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
0.143 |
-0.684 |
| 189 |
259570_at |
AT1G20440
|
AtCOR47, COR47, cold-regulated 47, RD17 |
0.143 |
-0.335 |
| 190 |
246288_at |
AT1G31850
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.142 |
0.353 |
| 191 |
245427_at |
AT4G17550
|
AtG3Pp4, glycerol-3-phosphate permease 4, G3Pp4, glycerol-3-phosphate permease 4 |
0.141 |
0.546 |
| 192 |
265694_at |
AT2G24440
|
[selenium binding] |
0.141 |
0.674 |
| 193 |
262743_at |
AT1G29020
|
[Calcium-binding EF-hand family protein] |
0.141 |
-0.864 |
| 194 |
252043_at |
AT3G52390
|
[TatD related DNase] |
0.141 |
0.061 |
| 195 |
264369_at |
AT1G70430
|
[Protein kinase superfamily protein] |
0.141 |
0.016 |
| 196 |
254202_at |
AT4G24140
|
[alpha/beta-Hydrolases superfamily protein] |
0.141 |
-0.349 |
| 197 |
262491_at |
AT1G21650
|
SECA2 |
0.141 |
-0.423 |
| 198 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
0.141 |
-0.109 |
| 199 |
248036_at |
AT5G55920
|
OLI2, OLIGOCELLULA 2 |
0.140 |
0.719 |
| 200 |
245807_at |
AT1G46768
|
RAP2.1, related to AP2 1 |
0.140 |
-0.908 |
| 201 |
250802_at |
AT5G04970
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.140 |
-0.633 |
| 202 |
267287_at |
AT2G23630
|
sks16, SKU5 similar 16 |
0.140 |
-0.087 |
| 203 |
260100_at |
AT1G73177
|
APC13, anaphase-promoting complex 13, BNS, BONSAI |
0.140 |
-0.063 |
| 204 |
257188_at |
AT3G13150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.139 |
0.777 |
| 205 |
250070_at |
AT5G17980
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.139 |
1.000 |
| 206 |
252391_at |
AT3G47860
|
CHL, chloroplastic lipocalin |
0.138 |
0.454 |
| 207 |
262892_at |
AT1G79440
|
ALDH5F1, aldehyde dehydrogenase 5F1, ENF1, ENLARGED FIL EXPRESSION DOMAIN 1, SSADH, SUCCINIC SEMIALDEHYDE DEHYDROGENASE, SSADH1, SUCCINIC SEMIALDEHYDE DEHYDROGENASE 1 |
0.138 |
-0.377 |
| 208 |
258868_at |
AT3G03110
|
CRM1B, XPO1B, exportin 1B |
0.138 |
0.643 |
| 209 |
267420_at |
AT2G35030
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.138 |
0.330 |
| 210 |
264535_at |
AT1G55690
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.138 |
0.235 |
| 211 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
0.137 |
1.428 |
| 212 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-0.917 |
-0.461 |
| 213 |
246001_at |
AT5G20790
|
unknown |
-0.911 |
-0.020 |
| 214 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-0.769 |
-0.334 |
| 215 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
-0.734 |
-0.545 |
| 216 |
266766_at |
AT2G46880
|
ATPAP14, PAP14, purple acid phosphatase 14 |
-0.699 |
-0.726 |
| 217 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.696 |
0.773 |
| 218 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
-0.686 |
-0.888 |
| 219 |
260668_at |
AT1G19530
|
unknown |
-0.659 |
-0.327 |
| 220 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
-0.628 |
-0.009 |
| 221 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.582 |
0.008 |
| 222 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.557 |
0.087 |
| 223 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.548 |
-0.295 |
| 224 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.508 |
-0.834 |
| 225 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.497 |
-0.859 |
| 226 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
-0.496 |
0.206 |
| 227 |
261814_at |
AT1G08310
|
[alpha/beta-Hydrolases superfamily protein] |
-0.491 |
-1.009 |
| 228 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.475 |
-0.480 |
| 229 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.461 |
-1.097 |
| 230 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
-0.438 |
0.150 |
| 231 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.436 |
-0.529 |
| 232 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
-0.427 |
-0.855 |
| 233 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.423 |
-0.395 |
| 234 |
258887_at |
AT3G05630
|
PDLZ2, PHOSPHOLIPASE D ZETA 2, PLDP2, phospholipase D P2, PLDZETA2, PHOSPHOLIPASE D ZETA 2 |
-0.423 |
-0.161 |
| 235 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.422 |
0.166 |
| 236 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
-0.422 |
-0.475 |
| 237 |
254111_at |
AT4G24890
|
ATPAP24, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 24, PAP24, purple acid phosphatase 24 |
-0.422 |
-1.021 |
| 238 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
-0.415 |
0.717 |
| 239 |
249614_at |
AT5G37300
|
WSD1 |
-0.410 |
0.806 |
| 240 |
252730_at |
AT3G43110
|
unknown |
-0.396 |
-0.207 |
| 241 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.387 |
-0.732 |
| 242 |
252520_at |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
-0.386 |
-0.923 |
| 243 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
-0.383 |
-0.182 |
| 244 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.377 |
-0.730 |
| 245 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
-0.375 |
0.096 |
| 246 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.374 |
0.622 |
| 247 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
-0.374 |
-0.869 |
| 248 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
-0.374 |
-0.199 |
| 249 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.364 |
-0.283 |
| 250 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.361 |
-0.767 |
| 251 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.358 |
-0.302 |
| 252 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
-0.351 |
-0.261 |
| 253 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.351 |
0.033 |
| 254 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
-0.340 |
-0.962 |
| 255 |
259015_at |
AT3G07350
|
unknown |
-0.336 |
-0.313 |
| 256 |
264774_at |
AT1G22890
|
unknown |
-0.334 |
-1.090 |
| 257 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.332 |
0.991 |
| 258 |
259982_at |
AT1G76410
|
ATL8 |
-0.332 |
-0.247 |
| 259 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
-0.331 |
0.042 |
| 260 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
-0.326 |
0.068 |
| 261 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
-0.326 |
-0.031 |
| 262 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
-0.323 |
-0.299 |
| 263 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.320 |
0.521 |
| 264 |
252073_at |
AT3G51750
|
unknown |
-0.316 |
-1.123 |
| 265 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.314 |
-1.099 |
| 266 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.313 |
-0.963 |
| 267 |
249932_at |
AT5G22390
|
unknown |
-0.311 |
-0.017 |
| 268 |
258225_at |
AT3G15630
|
unknown |
-0.308 |
1.374 |
| 269 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.308 |
-0.063 |
| 270 |
266278_at |
AT2G29300
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.305 |
0.365 |
| 271 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-0.305 |
0.938 |
| 272 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
-0.303 |
-0.561 |
| 273 |
248509_at |
AT5G50335
|
unknown |
-0.303 |
-0.779 |
| 274 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.301 |
0.859 |
| 275 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
-0.301 |
-1.379 |
| 276 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
-0.299 |
-0.512 |
| 277 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.299 |
-0.582 |
| 278 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
-0.298 |
-1.090 |
| 279 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.297 |
0.773 |
| 280 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
-0.297 |
-0.355 |
| 281 |
258939_at |
AT3G10020
|
unknown |
-0.293 |
0.327 |
| 282 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.293 |
-0.475 |
| 283 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
-0.293 |
-0.037 |
| 284 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.292 |
0.867 |
| 285 |
265387_at |
AT2G20670
|
unknown |
-0.291 |
0.246 |
| 286 |
257051_at |
AT3G15270
|
SPL5, squamosa promoter binding protein-like 5 |
-0.291 |
0.192 |
| 287 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-0.290 |
0.666 |
| 288 |
260012_at |
AT1G67865
|
unknown |
-0.289 |
-0.437 |
| 289 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.288 |
0.933 |
| 290 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.282 |
0.829 |
| 291 |
258421_at |
AT3G16690
|
AtSWEET16, SWEET16 |
-0.281 |
-0.727 |
| 292 |
250485_at |
AT5G09990
|
PROPEP5, elicitor peptide 5 precursor |
-0.280 |
-1.118 |
| 293 |
250327_at |
AT5G12050
|
unknown |
-0.280 |
-0.615 |
| 294 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
-0.279 |
0.556 |
| 295 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.278 |
0.686 |
| 296 |
256014_at |
AT1G19200
|
unknown |
-0.277 |
-0.357 |
| 297 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.277 |
-1.339 |
| 298 |
260075_at |
AT1G73700
|
[MATE efflux family protein] |
-0.277 |
-1.074 |
| 299 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
-0.274 |
-0.400 |
| 300 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.273 |
0.964 |
| 301 |
254683_at |
AT4G13800
|
unknown |
-0.272 |
-1.037 |
| 302 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
-0.271 |
0.440 |
| 303 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
-0.271 |
-0.593 |
| 304 |
264238_at |
AT1G54740
|
unknown |
-0.270 |
-1.118 |
| 305 |
247358_at |
AT5G63580
|
ATFLS2, FLS2, flavonol synthase 2 |
-0.269 |
-0.529 |
| 306 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.268 |
0.243 |
| 307 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
-0.268 |
-0.891 |
| 308 |
258570_at |
AT3G04530
|
ATPPCK2, PEPCK2, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 2, PPCK2, phosphoenolpyruvate carboxylase kinase 2 |
-0.267 |
-0.763 |
| 309 |
249791_at |
AT5G23810
|
AAP7, amino acid permease 7 |
-0.267 |
0.434 |
| 310 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.265 |
-0.179 |
| 311 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
-0.265 |
-0.359 |
| 312 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.263 |
0.042 |
| 313 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
-0.263 |
0.164 |
| 314 |
246783_at |
AT5G27360
|
SFP2 |
-0.263 |
0.308 |
| 315 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
-0.263 |
0.516 |
| 316 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.262 |
-0.424 |
| 317 |
255028_at |
AT4G09890
|
unknown |
-0.262 |
-0.593 |
| 318 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.260 |
-0.993 |
| 319 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
-0.260 |
0.085 |
| 320 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.255 |
-0.907 |
| 321 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
-0.253 |
-0.160 |
| 322 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
-0.252 |
-0.692 |
| 323 |
250881_at |
AT5G04080
|
unknown |
-0.252 |
-1.177 |
| 324 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.251 |
-0.199 |
| 325 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.249 |
-1.201 |
| 326 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
-0.248 |
-0.930 |
| 327 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-0.244 |
0.226 |
| 328 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
-0.241 |
-0.380 |
| 329 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.240 |
-0.759 |
| 330 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.239 |
0.865 |
| 331 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
-0.238 |
-0.516 |
| 332 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.237 |
-0.918 |
| 333 |
254240_at |
AT4G23496
|
SP1L5, SPIRAL1-like5 |
-0.237 |
-0.547 |
| 334 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.237 |
-0.827 |
| 335 |
250478_at |
AT5G10250
|
DOT3, DEFECTIVELY ORGANIZED TRIBUTARIES 3 |
-0.236 |
-1.026 |
| 336 |
247878_at |
AT5G57760
|
unknown |
-0.235 |
-1.063 |
| 337 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.233 |
-0.481 |
| 338 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
-0.229 |
1.309 |
| 339 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.229 |
-0.895 |
| 340 |
261224_at |
AT1G20160
|
ATSBT5.2 |
-0.227 |
-0.920 |
| 341 |
264580_at |
AT1G05340
|
unknown |
-0.227 |
-1.243 |
| 342 |
254638_at |
AT4G18740
|
[Rho termination factor] |
-0.224 |
-0.712 |
| 343 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.224 |
-0.022 |
| 344 |
249546_at |
AT5G38150
|
PMI15, plastid movement impaired 15 |
-0.223 |
-0.666 |
| 345 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
-0.222 |
-0.982 |
| 346 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
-0.222 |
0.101 |
| 347 |
266712_at |
AT2G46750
|
AtGulLO2, GulLO2, L -gulono-1,4-lactone ( L -GulL) oxidase 2 |
-0.221 |
-0.802 |
| 348 |
256757_at |
AT3G25620
|
ABCG21, ATP-binding cassette G21 |
-0.220 |
-0.490 |
| 349 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
-0.220 |
-0.219 |
| 350 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
-0.219 |
-0.082 |
| 351 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.219 |
0.449 |
| 352 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.219 |
-0.652 |
| 353 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
-0.217 |
-0.021 |
| 354 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.217 |
-0.874 |
| 355 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.217 |
-0.186 |
| 356 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.216 |
1.047 |
| 357 |
252057_at |
AT3G52480
|
unknown |
-0.216 |
-0.753 |
| 358 |
267089_at |
AT2G38300
|
[myb-like HTH transcriptional regulator family protein] |
-0.216 |
-1.530 |
| 359 |
251480_at |
AT3G59710
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.215 |
-0.842 |
| 360 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.215 |
-1.116 |
| 361 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
-0.215 |
-0.820 |
| 362 |
257672_at |
AT3G20300
|
unknown |
-0.213 |
-0.196 |
| 363 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
-0.213 |
-0.563 |
| 364 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.213 |
-1.457 |
| 365 |
262549_at |
AT1G31290
|
AGO3, ARGONAUTE 3 |
-0.212 |
0.299 |
| 366 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.212 |
0.601 |
| 367 |
257288_at |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
-0.212 |
-1.361 |
| 368 |
256672_at |
AT3G52310
|
ABCG27, ATP-binding cassette G27 |
-0.209 |
-0.024 |
| 369 |
259209_at |
AT3G09160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.208 |
-0.050 |
| 370 |
258935_at |
AT3G10120
|
unknown |
-0.207 |
-1.408 |
| 371 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.206 |
-0.950 |
| 372 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-0.205 |
0.694 |
| 373 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
-0.205 |
0.379 |
| 374 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.205 |
0.725 |
| 375 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-0.204 |
-0.014 |
| 376 |
264887_at |
AT1G23120
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.203 |
-0.912 |
| 377 |
263953_at |
AT2G36050
|
ATOFP15, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 15, OFP15, ovate family protein 15 |
-0.203 |
0.163 |
| 378 |
251750_at |
AT3G55710
|
[UDP-Glycosyltransferase superfamily protein] |
-0.203 |
-0.083 |
| 379 |
258782_at |
AT3G11750
|
FOLB1 |
-0.202 |
-1.187 |
| 380 |
265494_at |
AT2G15680
|
AtCML30, CML30, calmodulin-like 30 |
-0.202 |
-0.333 |
| 381 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
-0.202 |
0.614 |
| 382 |
251141_at |
AT5G01075
|
[Glycosyl hydrolase family 35 protein] |
-0.201 |
0.925 |
| 383 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.201 |
0.042 |
| 384 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
-0.201 |
-1.149 |
| 385 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.201 |
-0.269 |
| 386 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.200 |
-0.003 |
| 387 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.200 |
-0.093 |
| 388 |
263851_at |
AT2G04460
|
unknown |
-0.200 |
0.239 |
| 389 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.199 |
0.050 |
| 390 |
265769_at |
AT2G48090
|
unknown |
-0.198 |
-0.773 |
| 391 |
251235_at |
AT3G62860
|
[alpha/beta-Hydrolases superfamily protein] |
-0.195 |
-0.670 |
| 392 |
263032_at |
AT1G23850
|
unknown |
-0.195 |
-0.035 |
| 393 |
259001_at |
AT3G01960
|
unknown |
-0.195 |
-0.260 |
| 394 |
248801_at |
AT5G47370
|
HAT2 |
-0.195 |
0.153 |
| 395 |
260735_at |
AT1G17610
|
CHS1, CHILLING SENSITIVE 1 |
-0.194 |
-0.886 |
| 396 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.194 |
0.048 |
| 397 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
-0.194 |
0.542 |
| 398 |
249765_at |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
-0.193 |
-0.645 |
| 399 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
-0.193 |
0.035 |
| 400 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.193 |
-0.189 |
| 401 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
-0.193 |
0.407 |
| 402 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.192 |
-0.723 |
| 403 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
-0.192 |
-0.269 |
| 404 |
258189_at |
AT3G17860
|
JAI3, JASMONATE-INSENSITIVE 3, JAZ3, jasmonate-zim-domain protein 3, TIFY6B |
-0.192 |
-0.461 |
| 405 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.192 |
0.348 |
| 406 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
-0.189 |
0.589 |
| 407 |
253519_at |
AT4G31240
|
AtNRX2, NRX2, nucleoredoxin 2 |
-0.189 |
-0.252 |
| 408 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
-0.188 |
-0.569 |
| 409 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
-0.187 |
-0.307 |
| 410 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.187 |
-0.651 |
| 411 |
254175_at |
AT4G24050
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.186 |
0.911 |
| 412 |
252412_at |
AT3G47295
|
unknown |
-0.186 |
0.229 |
| 413 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
-0.185 |
0.211 |
| 414 |
254522_at |
AT4G19980
|
unknown |
-0.185 |
-0.662 |
| 415 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
-0.184 |
-0.996 |
| 416 |
261222_at |
AT1G20120
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.184 |
-0.474 |
| 417 |
258468_at |
AT3G06070
|
unknown |
-0.183 |
-1.099 |
| 418 |
251010_at |
AT5G02550
|
unknown |
-0.182 |
0.801 |
| 419 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
-0.182 |
-0.181 |
| 420 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
-0.181 |
-0.290 |
| 421 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
-0.181 |
1.010 |