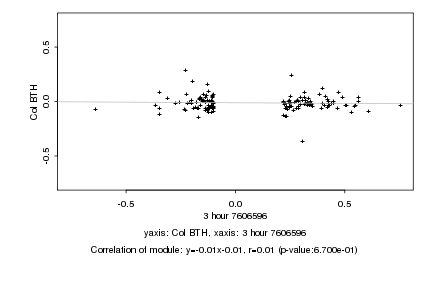
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
3 hour 7606596 |
Log2 signal ratio
Col BTH |
| 1 |
262563_at |
AT1G34210
|
ATSERK2, SERK2, somatic embryogenesis receptor-like kinase 2 |
0.753 |
-0.029 |
| 2 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
0.605 |
-0.084 |
| 3 |
253142_at |
AT4G35520
|
ATMLH3, MLH3, MUTL protein homolog 3 |
0.562 |
0.004 |
| 4 |
258716_at |
AT3G09700
|
[Chaperone DnaJ-domain superfamily protein] |
0.560 |
0.039 |
| 5 |
258869_at |
AT3G03090
|
AtVGT1, vacuolar glucose transporter 1, VGT1, vacuolar glucose transporter 1 |
0.545 |
-0.034 |
| 6 |
248439_at |
AT5G51250
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.542 |
-0.038 |
| 7 |
267013_at |
AT2G39180
|
ATCRR2, CCR2, CRINKLY4 related 2 |
0.527 |
-0.096 |
| 8 |
246857_at |
AT5G25920
|
unknown |
0.507 |
-0.028 |
| 9 |
248427_at |
AT5G51750
|
ATSBT1.3, subtilase 1.3, SBT1.3, subtilase 1.3 |
0.501 |
-0.031 |
| 10 |
263383_at |
AT2G40120
|
[Protein kinase superfamily protein] |
0.485 |
0.040 |
| 11 |
250786_at |
AT5G05540
|
SDN2, small RNA degrading nuclease 2 |
0.467 |
0.089 |
| 12 |
262786_at |
AT1G10740
|
[alpha/beta-Hydrolases superfamily protein] |
0.464 |
-0.060 |
| 13 |
267212_at |
AT2G44060
|
[Late embryogenesis abundant protein, group 2] |
0.448 |
0.006 |
| 14 |
257269_at |
AT3G17440
|
ATNPSN13, NPSN13, novel plant snare 13 |
0.444 |
-0.014 |
| 15 |
266202_at |
AT2G02400
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.436 |
-0.017 |
| 16 |
257265_at |
AT3G14980
|
IDM1, increased DNA methylation 1, ROS4, REPRESSOR OF SILENCING 4 |
0.429 |
-0.032 |
| 17 |
259861_at |
AT1G80620
|
[S15/NS1, RNA-binding protein] |
0.425 |
0.005 |
| 18 |
260181_at |
AT1G70710
|
ATGH9B1, glycosyl hydrolase 9B1, CEL1, CELLULASE 1, GH9B1, glycosyl hydrolase 9B1 |
0.423 |
-0.012 |
| 19 |
258008_at |
AT3G19430
|
[late embryogenesis abundant protein-related / LEA protein-related] |
0.421 |
-0.032 |
| 20 |
250414_at |
AT5G11180
|
ATGLR2.6, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 2.6, GLR2.6, glutamate receptor 2.6 |
0.418 |
-0.054 |
| 21 |
246295_at |
AT3G56690
|
CIP111, Cam interacting protein 111 |
0.418 |
0.022 |
| 22 |
265630_at |
AT2G27350
|
OTLD1, otubain-like deubiquitinase 1 |
0.411 |
0.047 |
| 23 |
256546_at |
AT3G14820
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.406 |
-0.029 |
| 24 |
264227_at |
AT1G67500
|
ATREV3, recovery protein 3, REV3, recovery protein 3 |
0.397 |
-0.009 |
| 25 |
249409_at |
AT5G40340
|
[Tudor/PWWP/MBT superfamily protein] |
0.394 |
0.124 |
| 26 |
264426_at |
AT1G61760
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.392 |
-0.058 |
| 27 |
262664_at |
AT1G13970
|
unknown |
0.381 |
0.067 |
| 28 |
250637_at |
AT5G07530
|
ATGRP-7, ARABIDOPSIS THALIANA GLYCINE RICH PROTEIN 7, ATGRP17, ARABIDOPSIS THALIANA GLYCINE RICH PROTEIN 17, GRP17, glycine rich protein 17 |
0.352 |
-0.044 |
| 29 |
247743_at |
AT5G59010
|
BSK5, brassinosteroid-signaling kinase 5 |
0.346 |
-0.010 |
| 30 |
246938_at |
AT5G25380
|
CYCA2;1, cyclin a2;1 |
0.346 |
-0.019 |
| 31 |
265472_at |
AT2G15580
|
[RING/U-box superfamily protein] |
0.342 |
0.002 |
| 32 |
267250_at |
AT2G23060
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.335 |
-0.023 |
| 33 |
263123_at |
AT1G78500
|
PEN6, PENTACYCLIC TRITERPENE SYNTHASE 6 |
0.335 |
-0.028 |
| 34 |
262754_at |
AT1G16350
|
[Aldolase-type TIM barrel family protein] |
0.331 |
-0.016 |
| 35 |
266206_at |
AT2G27730
|
[copper ion binding] |
0.330 |
0.032 |
| 36 |
266916_at |
AT2G45860
|
unknown |
0.327 |
-0.034 |
| 37 |
250762_at |
AT5G05990
|
[Mitochondrial glycoprotein family protein] |
0.326 |
-0.008 |
| 38 |
264406_at |
AT1G10290
|
ADL6, dynamin-like protein 6, DRP2A, DYNAMIN-RELATED PROTEIN 2A |
0.317 |
0.010 |
| 39 |
249900_at |
AT5G22640
|
emb1211, embryo defective 1211, TIC100, TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 100 |
0.317 |
-0.010 |
| 40 |
261212_at |
AT1G12880
|
atnudt12, nudix hydrolase homolog 12, NUDT12, nudix hydrolase homolog 12 |
0.314 |
0.090 |
| 41 |
252264_at |
AT3G49590
|
ATG13, autophagy-related 13 |
0.312 |
0.039 |
| 42 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.312 |
-0.021 |
| 43 |
253232_at |
AT4G34340
|
TAF8, TBP-associated factor 8 |
0.306 |
0.016 |
| 44 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.303 |
-0.366 |
| 45 |
251588_at |
AT3G58090
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.297 |
0.045 |
| 46 |
266812_at |
AT2G44830
|
[Protein kinase superfamily protein] |
0.293 |
-0.024 |
| 47 |
259189_at |
AT3G01700
|
AGP11, arabinogalactan protein 11, ATAGP11, ARABINOGALACTAN PROTEIN 11 |
0.288 |
-0.040 |
| 48 |
248819_at |
AT5G47050
|
[SBP (S-ribonuclease binding protein) family protein] |
0.288 |
0.009 |
| 49 |
248454_at |
AT5G51350
|
MOL1, MORE LATERAL GROWTH1 |
0.284 |
-0.058 |
| 50 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
0.281 |
0.018 |
| 51 |
249897_at |
AT5G22550
|
unknown |
0.280 |
0.012 |
| 52 |
247141_at |
AT5G65560
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.279 |
0.007 |
| 53 |
263757_at |
AT2G21430
|
[Papain family cysteine protease] |
0.275 |
-0.062 |
| 54 |
260632_at |
AT1G62360
|
BUM, BUMBERSHOOT, BUM1, BUMBERSHOOT 1, SHL, SHOOTLESS, STM, SHOOT MERISTEMLESS, WAM, WALDMEISTER, WAM1, WALDMEISTER 1 |
0.266 |
-0.007 |
| 55 |
244939_at |
ATCG00065
|
RPS12, RIBOSOMAL PROTEIN S12, RPS12A, ribosomal protein S12A |
0.264 |
-0.078 |
| 56 |
256778_at |
AT3G13782
|
NAP1;4, nucleosome assembly protein1;4, NFA04, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A 04, NFA4, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A 4 |
0.255 |
-0.044 |
| 57 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.252 |
0.245 |
| 58 |
263362_at |
AT2G03880
|
REME1, required for efficiency of mitochondrial editing 1 |
0.251 |
0.051 |
| 59 |
255522_at |
AT4G02260
|
AT-RSH1, RELA-SPOT HOMOLOG 1, ATRSH1, RELA/SPOT HOMOLOG 1, RSH1, RELA/SPOT homolog 1 |
0.250 |
-0.043 |
| 60 |
266576_at |
AT2G23940
|
unknown |
0.248 |
-0.008 |
| 61 |
262323_at |
AT1G64190
|
[6-phosphogluconate dehydrogenase family protein] |
0.247 |
-0.014 |
| 62 |
253089_at |
AT4G36290
|
AtMORC1, CRT1, compromised recognition of TCV 1, MORC1, Microchidia 1 |
0.247 |
0.014 |
| 63 |
258549_at |
AT3G06930
|
ATPRMT4B, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 4B, PRMT4B, protein arginine methyltransferase 4B |
0.243 |
-0.047 |
| 64 |
256912_at |
AT3G23850
|
unknown |
0.243 |
-0.037 |
| 65 |
267581_at |
AT2G41980
|
[Protein with RING/U-box and TRAF-like domains] |
0.241 |
0.004 |
| 66 |
252781_at |
AT3G42950
|
[Pectin lyase-like superfamily protein] |
0.236 |
-0.068 |
| 67 |
250610_at |
AT5G07550
|
ATGRP19, GRP19, glycine-rich protein 19 |
0.233 |
-0.042 |
| 68 |
257571_at |
AT3G16870
|
GATA17, GATA transcription factor 17 |
0.230 |
-0.135 |
| 69 |
253464_at |
AT4G32030
|
unknown |
0.228 |
-0.017 |
| 70 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.227 |
-0.130 |
| 71 |
259435_at |
AT1G01450
|
[Protein kinase superfamily protein] |
0.226 |
-0.062 |
| 72 |
266549_at |
AT2G35150
|
EXL7, EXORDIUM LIKE 7 |
0.220 |
-0.021 |
| 73 |
257208_at |
AT3G14910
|
unknown |
0.219 |
0.007 |
| 74 |
258653_at |
AT3G09870
|
SAUR48, SMALL AUXIN UPREGULATED RNA 48 |
0.218 |
-0.124 |
| 75 |
254443_at |
AT4G21070
|
ATBRCA1, ARABIDOPSIS THALIANA BREAST CANCER SUSCEPTIBILITY1, BRCA1, breast cancer susceptibility1 |
0.217 |
-0.004 |
| 76 |
266022_at |
AT2G05920
|
[Subtilase family protein] |
0.216 |
-0.008 |
| 77 |
261301_at |
AT1G48570
|
[zinc finger (Ran-binding) family protein] |
-0.644 |
-0.066 |
| 78 |
260895_at |
AT1G29250
|
[Alba DNA/RNA-binding protein] |
-0.370 |
-0.032 |
| 79 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
-0.351 |
-0.118 |
| 80 |
249044_at |
AT5G44370
|
PHT4;6, phosphate transporter 4;6 |
-0.349 |
-0.060 |
| 81 |
248279_at |
AT5G52910
|
ATIM, TIMELESS |
-0.349 |
0.088 |
| 82 |
264028_at |
AT2G03680
|
SKU6, SPR1, SPIRAL1 |
-0.314 |
0.036 |
| 83 |
263691_at |
AT1G26880
|
[Ribosomal protein L34e superfamily protein] |
-0.278 |
-0.016 |
| 84 |
254310_at |
AT4G22430
|
[F-box family protein] |
-0.258 |
-0.004 |
| 85 |
261799_at |
AT1G30473
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.235 |
-0.070 |
| 86 |
247087_at |
AT5G66330
|
[Leucine-rich repeat (LRR) family protein] |
-0.232 |
-0.074 |
| 87 |
245401_at |
AT4G17670
|
unknown |
-0.231 |
0.294 |
| 88 |
262255_at |
AT1G53790
|
[F-box and associated interaction domains-containing protein] |
-0.225 |
0.067 |
| 89 |
262169_at |
AT1G74860
|
unknown |
-0.220 |
-0.013 |
| 90 |
250162_at |
AT5G15250
|
ATFTSH6, FTSH6, FTSH protease 6 |
-0.211 |
-0.001 |
| 91 |
256628_at |
AT3G20000
|
TOM40, translocase of the outer mitochondrial membrane 40 |
-0.202 |
-0.015 |
| 92 |
261887_at |
AT1G80780
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
-0.202 |
0.011 |
| 93 |
250775_at |
AT5G05460
|
AtENGase85A, ENGase85A, Endo-beta-N-acetyglucosaminidase 85A |
-0.200 |
0.186 |
| 94 |
255393_at |
AT4G03690
|
unknown |
-0.196 |
-0.061 |
| 95 |
249054_at |
AT5G44450
|
[methyltransferases] |
-0.187 |
-0.046 |
| 96 |
261563_at |
AT1G01630
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.181 |
-0.002 |
| 97 |
258656_at |
AT3G09900
|
ATRAB8E, ATRABE1E, RAB GTPase homolog E1E, RABE1e, RAB GTPase homolog E1E |
-0.179 |
-0.005 |
| 98 |
248446_at |
AT5G51140
|
[Pseudouridine synthase family protein] |
-0.175 |
-0.057 |
| 99 |
255142_at |
AT4G08390
|
SAPX, stromal ascorbate peroxidase |
-0.173 |
-0.063 |
| 100 |
254512_at |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.172 |
-0.141 |
| 101 |
251257_at |
AT3G62290
|
ARFA1E, ADP-ribosylation factor A1E, ATARFA1E, ADP-ribosylation factor A1E |
-0.166 |
0.033 |
| 102 |
260489_at |
AT1G51610
|
[Cation efflux family protein] |
-0.165 |
0.021 |
| 103 |
249322_at |
AT5G40930
|
TOM20-4, translocase of outer membrane 20-4 |
-0.164 |
0.040 |
| 104 |
263661_at |
AT1G04290
|
[Thioesterase superfamily protein] |
-0.163 |
-0.030 |
| 105 |
257159_at |
AT3G24400
|
AtPERK2, proline-rich extensin-like receptor kinase 2, PERK2, proline-rich extensin-like receptor kinase 2 |
-0.157 |
0.023 |
| 106 |
246738_at |
AT5G27740
|
EMB161, EMBRYO DEFECTIVE 161, EMB251, EMBRYO DEFECTIVE 251, EMB2775, EMBRYO DEFECTIVE 2775, RFC3, replication factor C 3 |
-0.155 |
0.002 |
| 107 |
263186_at |
AT1G36110
|
[copia-like retrotransposon family, has a 2.2e-59 P-value blast match to gb|AAO73521.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-0.153 |
0.010 |
| 108 |
246584_at |
AT5G14730
|
unknown |
-0.150 |
0.006 |
| 109 |
257541_at |
AT3G25950
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
-0.150 |
0.010 |
| 110 |
257636_at |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
-0.147 |
0.068 |
| 111 |
249962_at |
AT5G18990
|
[Pectin lyase-like superfamily protein] |
-0.144 |
0.002 |
| 112 |
264096_at |
AT1G78995
|
unknown |
-0.139 |
-0.081 |
| 113 |
249898_at |
AT5G22510
|
A/N-InvE, alkaline/neutral invertase E, At-A/N-InvE, Arabidopsis alkaline/neutral invertase E, INV-E, alkaline/neutral invertase E |
-0.139 |
-0.007 |
| 114 |
260992_at |
AT1G12150
|
unknown |
-0.139 |
0.042 |
| 115 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.137 |
-0.062 |
| 116 |
251763_at |
AT3G55730
|
AtMYB109, myb domain protein 109, MYB109, myb domain protein 109 |
-0.133 |
0.064 |
| 117 |
267079_at |
AT2G41200
|
unknown |
-0.132 |
-0.072 |
| 118 |
260434_at |
AT1G68330
|
unknown |
-0.129 |
-0.080 |
| 119 |
259911_at |
AT1G72680
|
ATCAD1, CINNAMYL ALCOHOL DEHYDROGENASE 1, CAD1, cinnamyl-alcohol dehydrogenase |
-0.128 |
0.164 |
| 120 |
262747_at |
AT1G28540
|
unknown |
-0.128 |
0.011 |
| 121 |
263081_at |
AT2G27220
|
BLH5, BEL1-like homeodomain 5 |
-0.127 |
-0.033 |
| 122 |
260119_at |
AT1G33930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.126 |
-0.094 |
| 123 |
260242_at |
AT1G63650
|
AtEGL3, ATMYC-2, EGL1, EGL3, ENHANCER OF GLABRA 3 |
-0.126 |
-0.048 |
| 124 |
247418_at |
AT5G63030
|
GRXC1, glutaredoxin C1 |
-0.125 |
0.096 |
| 125 |
257258_at |
AT3G22040
|
unknown |
-0.121 |
-0.058 |
| 126 |
266913_at |
AT2G45890
|
ATROPGEF4, RHO GUANYL-NUCLEOTIDE EXCHANGE FACTOR 4, GEF4, guanine exchange factor 4, RHS11, ROOT HAIR SPECIFIC 11, ROPGEF4, ROP (rho of plants) guanine nucleotide exchange factor 4 |
-0.120 |
0.004 |
| 127 |
249990_at |
AT5G18540
|
unknown |
-0.120 |
0.003 |
| 128 |
251831_at |
AT3G55140
|
[Pectin lyase-like superfamily protein] |
-0.119 |
-0.049 |
| 129 |
267338_at |
AT2G39990
|
AteIF3f, Arabidopsis thaliana eukaryotic translation initiation factor 3 subunit F, EIF2, eukaryotic translation initiation factor 2, eIF3F, eukaryotic translation initiation factor 3 subunit F |
-0.114 |
0.010 |
| 130 |
259864_at |
AT1G72800
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.114 |
-0.099 |
| 131 |
259305_at |
AT3G05070
|
unknown |
-0.113 |
-0.001 |
| 132 |
255162_at |
AT4G07840
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:BAA22288 pol polyprotein (Ty1_Copia-element) (Oryza australiensis)GB:BAA22288 polyprotein (Ty1_Copia-element) (Oryza australiensis)gi|2443320|dbj|BAA22288.1| polyprotein (RIRE1) (Oryza australiensis) (Ty1_Copia-element)] |
-0.112 |
-0.037 |
| 133 |
251608_at |
AT3G57860
|
GIG1, GIGAS CELL 1, OSD1, OMISSION OF SECOND DIVISION, UVI4-LIKE, UV-B-insensitive 4-like |
-0.112 |
-0.032 |
| 134 |
260043_at |
AT1G41770
|
unknown |
-0.110 |
-0.038 |
| 135 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
-0.109 |
-0.050 |
| 136 |
246536_at |
AT5G15920
|
EMB2782, EMBRYO DEFECTIVE 2782, SMC5, structural maintenance of chromosomes 5 |
-0.108 |
-0.008 |
| 137 |
263772_at |
AT2G06255
|
ELF4-L3, ELF4-like 3 |
-0.108 |
0.060 |
| 138 |
249592_at |
AT5G37890
|
[Protein with RING/U-box and TRAF-like domains] |
-0.108 |
-0.005 |
| 139 |
250607_at |
AT5G07370
|
ATIPK2A, INOSITOL POLYPHOSPHATE KINASE 2 ALPHA, IMPK, inositol multi-phosphate kinase, IPK2a, inositol polyphosphate kinase 2 alpha |
-0.107 |
0.049 |
| 140 |
253371_at |
AT4G33240
|
FAB1A, FORMS APLOID AND BINUCLEATE CELLS 1A |
-0.106 |
-0.040 |
| 141 |
252872_at |
AT4G40010
|
SNRK2-7, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-7, SNRK2.7, SNF1-related protein kinase 2.7, SRK2F |
-0.106 |
-0.043 |
| 142 |
255428_at |
AT4G03390
|
SRF3, STRUBBELIG-receptor family 3 |
-0.106 |
0.041 |
| 143 |
245832_at |
AT1G48850
|
EMB1144, embryo defective 1144 |
-0.105 |
0.004 |
| 144 |
267267_at |
AT2G02580
|
CYP71B9, cytochrome P450, family 71, subfamily B, polypeptide 9 |
-0.105 |
-0.060 |
| 145 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
-0.104 |
-0.056 |
| 146 |
260409_at |
AT1G69935
|
SHW1, SHORT HYPOCOTYL IN WHITE LIGHT1 |
-0.103 |
-0.006 |
| 147 |
252220_at |
AT3G49940
|
LBD38, LOB domain-containing protein 38 |
-0.103 |
0.068 |
| 148 |
259717_at |
AT1G61010
|
CPSF73-I, cleavage and polyadenylation specificity factor 73-I |
-0.103 |
-0.038 |
| 149 |
265188_at |
AT1G23800
|
ALDH2B, aldehyde dehydrogenase 2B, ALDH2B7, aldehyde dehydrogenase 2B7 |
-0.102 |
-0.053 |
| 150 |
248929_at |
AT5G46000
|
[Mannose-binding lectin superfamily protein] |
-0.102 |
-0.017 |
| 151 |
251093_at |
AT5G01360
|
TBL3, TRICHOME BIREFRINGENCE-LIKE 3 |
-0.102 |
-0.091 |