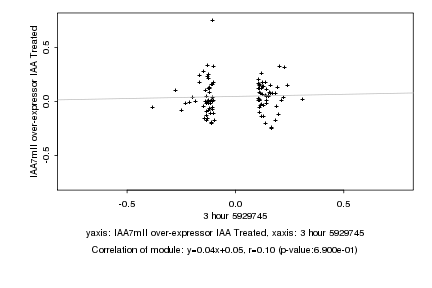
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
3 hour 5929745 |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
244939_at |
ATCG00065
|
RPS12, RIBOSOMAL PROTEIN S12, RPS12A, ribosomal protein S12A |
0.307 |
0.027 |
| 2 |
265177_at |
AT1G23640
|
unknown |
0.239 |
0.153 |
| 3 |
246426_at |
AT5G17430
|
BBM, BABY BOOM |
0.222 |
0.321 |
| 4 |
245012_at |
ATCG00440
|
NDHC |
0.220 |
0.043 |
| 5 |
258008_at |
AT3G19430
|
[late embryogenesis abundant protein-related / LEA protein-related] |
0.212 |
0.010 |
| 6 |
266691_at |
AT2G19890
|
unknown |
0.203 |
0.332 |
| 7 |
246391_at |
AT1G77350
|
unknown |
0.197 |
-0.114 |
| 8 |
252070_at |
AT3G51680
|
AtSDR2, SDR2, short-chain dehydrogenase/reductase 2 |
0.192 |
0.135 |
| 9 |
262142_at |
AT1G52640
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.186 |
-0.042 |
| 10 |
244996_at |
ATCG00160
|
RPS2, ribosomal protein S2 |
0.183 |
0.076 |
| 11 |
259861_at |
AT1G80620
|
[S15/NS1, RNA-binding protein] |
0.182 |
-0.168 |
| 12 |
244921_s_at |
ATMG01000
|
ORF114 |
0.169 |
0.081 |
| 13 |
247703_at |
AT5G59560
|
SRR1, SENSITIVITY TO RED LIGHT REDUCED 1 |
0.166 |
-0.234 |
| 14 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
0.163 |
-0.243 |
| 15 |
250289_at |
AT5G13190
|
AtGILP, GILP, GSH-induced LITAF domain protein |
0.159 |
0.080 |
| 16 |
252280_at |
AT3G49260
|
iqd21, IQ-domain 21 |
0.158 |
0.153 |
| 17 |
264406_at |
AT1G10290
|
ADL6, dynamin-like protein 6, DRP2A, DYNAMIN-RELATED PROTEIN 2A |
0.157 |
0.091 |
| 18 |
250110_at |
AT5G15350
|
AtENODL17, ENODL17, early nodulin-like protein 17 |
0.150 |
0.054 |
| 19 |
245019_at |
ATCG00530
|
YCF10 |
0.148 |
0.059 |
| 20 |
255124_at |
AT4G08560
|
APUM15, pumilio 15, PUM15, pumilio 15 |
0.143 |
-0.011 |
| 21 |
246901_at |
AT5G25630
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.142 |
0.051 |
| 22 |
256161_at |
AT1G30090
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.142 |
0.012 |
| 23 |
252264_at |
AT3G49590
|
ATG13, autophagy-related 13 |
0.140 |
0.120 |
| 24 |
259848_at |
AT1G72180
|
[Leucine-rich receptor-like protein kinase family protein] |
0.136 |
-0.203 |
| 25 |
266032_x_at |
AT2G05890
|
unknown |
0.136 |
0.056 |
| 26 |
249581_at |
AT5G37600
|
ATGLN1;1, ARABIDOPSIS GLUTAMINE SYNTHASE 1;1, ATGSR1, ARABIDOPSIS THALIANA GLUTAMINE SYNTHASE CLONE R1, GLN1;1, GLUTAMINE SYNTHASE 1;1, GSR 1, glutamine synthase clone R1 |
0.135 |
0.185 |
| 27 |
254671_at |
AT4G18410
|
[Mutator-like transposase family, has a 8.7e-30 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.127 |
0.141 |
| 28 |
261935_at |
AT1G22610
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.127 |
-0.029 |
| 29 |
245004_at |
ATCG00300
|
YCF9 |
0.126 |
-0.133 |
| 30 |
251086_at |
AT5G01450
|
APD2, ABERRANT POLLEN DEVELOPMENT 2 |
0.124 |
0.066 |
| 31 |
250091_at |
AT5G17340
|
[Putative membrane lipoprotein] |
0.122 |
0.182 |
| 32 |
246240_at |
AT4G37130
|
TCU1, TRANSCURVATA 1 |
0.121 |
0.140 |
| 33 |
266549_at |
AT2G35150
|
EXL7, EXORDIUM LIKE 7 |
0.118 |
0.266 |
| 34 |
253967_at |
AT4G26550
|
[Got1/Sft2-like vescicle transport protein family] |
0.117 |
-0.130 |
| 35 |
249094_at |
AT5G43890
|
SUPER1, SUPPRESSOR OF ER 1, YUC5, YUCCA5 |
0.117 |
0.127 |
| 36 |
251659_at |
AT3G57090
|
BIGYIN, FIS1A, FISSION 1A |
0.116 |
-0.020 |
| 37 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
0.113 |
0.079 |
| 38 |
262973_at |
AT1G75600
|
[Histone superfamily protein] |
0.112 |
-0.034 |
| 39 |
254405_at |
AT4G21350
|
B80, PUB8, plant U-box 8 |
0.111 |
-0.095 |
| 40 |
258185_at |
AT3G21580
|
ABCI12, ATP-binding cassette I12 |
0.111 |
-0.053 |
| 41 |
246007_at |
AT5G08410
|
FTRA2, ferredoxin/thioredoxin reductase subunit A (variable subunit) 2 |
0.111 |
0.155 |
| 42 |
267581_at |
AT2G41980
|
[Protein with RING/U-box and TRAF-like domains] |
0.111 |
0.013 |
| 43 |
261953_at |
AT1G64440
|
REB1, ROOT EPIDERMAL BULGER1, RHD1, ROOT HAIR DEFECTIVE 1, UGE4, UDP-GLUCOSE 4-EPIMERASE |
0.110 |
0.020 |
| 44 |
250839_at |
AT5G04640
|
AGL99, AGAMOUS-like 99 |
0.109 |
0.124 |
| 45 |
260763_at |
AT1G49220
|
[RING/U-box superfamily protein] |
0.108 |
0.159 |
| 46 |
264444_at |
AT1G27360
|
SPL11, squamosa promoter-like 11 |
0.108 |
0.087 |
| 47 |
265286_at |
AT2G20360
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.107 |
0.010 |
| 48 |
261064_at |
AT1G07510
|
ftsh10, FTSH protease 10 |
0.106 |
0.015 |
| 49 |
245020_at |
ATCG00540
|
PETA, photosynthetic electron transfer A |
0.105 |
0.122 |
| 50 |
266665_at |
AT2G29790
|
[Maternally expressed gene (MEG) family protein] |
0.105 |
0.173 |
| 51 |
247438_at |
AT5G62460
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.104 |
0.205 |
| 52 |
245008_at |
ATCG00360
|
YCF3 |
0.102 |
0.036 |
| 53 |
262377_at |
AT1G73110
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.384 |
-0.049 |
| 54 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
-0.280 |
0.105 |
| 55 |
264028_at |
AT2G03680
|
SKU6, SPR1, SPIRAL1 |
-0.252 |
-0.078 |
| 56 |
249837_at |
AT5G23480
|
[SWIB/MDM2 domain] |
-0.233 |
-0.015 |
| 57 |
266091_at |
AT2G37920
|
emb1513, embryo defective 1513 |
-0.215 |
-0.006 |
| 58 |
249322_at |
AT5G40930
|
TOM20-4, translocase of outer membrane 20-4 |
-0.199 |
0.045 |
| 59 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
-0.185 |
0.004 |
| 60 |
257120_at |
AT3G20200
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.169 |
0.246 |
| 61 |
264075_at |
AT2G10850
|
unknown |
-0.168 |
0.181 |
| 62 |
247030_at |
AT5G67210
|
IRX15-L, IRX15-LIKE |
-0.149 |
0.281 |
| 63 |
249145_at |
AT5G43280
|
ATDCI1, delta(3,5),delta(2,4)-dienoyl-CoA isomerase 1, DCI1, delta(3,5),delta(2,4)-dienoyl-CoA isomerase 1, DCI1, DELTA(3,5),DELTA(2,4)-DIENOYL-COA ISOMERASE 1 |
-0.149 |
-0.038 |
| 64 |
261563_at |
AT1G01630
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.145 |
-0.153 |
| 65 |
264096_at |
AT1G78995
|
unknown |
-0.140 |
0.104 |
| 66 |
249990_at |
AT5G18540
|
unknown |
-0.139 |
0.002 |
| 67 |
246738_at |
AT5G27740
|
EMB161, EMBRYO DEFECTIVE 161, EMB251, EMBRYO DEFECTIVE 251, EMB2775, EMBRYO DEFECTIVE 2775, RFC3, replication factor C 3 |
-0.138 |
-0.169 |
| 68 |
248279_at |
AT5G52910
|
ATIM, TIMELESS |
-0.138 |
-0.018 |
| 69 |
251669_at |
AT3G57180
|
BPG2, BRASSINAZOLE(BRZ) INSENSITIVE PALE GREEN 2 |
-0.137 |
0.051 |
| 70 |
251763_at |
AT3G55730
|
AtMYB109, myb domain protein 109, MYB109, myb domain protein 109 |
-0.135 |
-0.087 |
| 71 |
250775_at |
AT5G05460
|
AtENGase85A, ENGase85A, Endo-beta-N-acetyglucosaminidase 85A |
-0.135 |
-0.126 |
| 72 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.133 |
0.238 |
| 73 |
244908_at |
ATMG00720
|
ORF107D |
-0.133 |
0.335 |
| 74 |
250981_at |
AT5G03140
|
LecRK-VIII.2, L-type lectin receptor kinase VIII.2 |
-0.132 |
-0.151 |
| 75 |
266926_at |
AT2G46000
|
unknown |
-0.132 |
-0.000 |
| 76 |
246305_at |
AT3G51890
|
CLC3, clathrin light chain 3 |
-0.132 |
0.008 |
| 77 |
263754_at |
AT2G21510
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.127 |
0.253 |
| 78 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
-0.127 |
-0.012 |
| 79 |
260573_at |
AT2G47280
|
[Pectin lyase-like superfamily protein] |
-0.127 |
-0.074 |
| 80 |
252532_at |
AT3G46570
|
[Glycosyl hydrolase superfamily protein] |
-0.126 |
0.222 |
| 81 |
253858_at |
AT4G27600
|
NARA5, GENES NECESSARY FOR THE ACHIEVEMENT OF RUBISCO ACCUMULATION 5 |
-0.126 |
0.122 |
| 82 |
251279_at |
AT3G61800
|
unknown |
-0.125 |
0.010 |
| 83 |
252360_at |
AT3G48480
|
[Cysteine proteinases superfamily protein] |
-0.124 |
0.137 |
| 84 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
-0.124 |
0.090 |
| 85 |
248867_at |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
-0.123 |
0.125 |
| 86 |
260344_at |
AT1G69240
|
ATMES15, ARABIDOPSIS THALIANA METHYL ESTERASE 15, MES15, methyl esterase 15, RHS9, ROOT HAIR SPECIFIC 9 |
-0.121 |
-0.064 |
| 87 |
245310_at |
AT4G13950
|
RALFL31, ralf-like 31 |
-0.118 |
-0.108 |
| 88 |
265849_at |
AT2G35736
|
unknown |
-0.117 |
0.007 |
| 89 |
267338_at |
AT2G39990
|
AteIF3f, Arabidopsis thaliana eukaryotic translation initiation factor 3 subunit F, EIF2, eukaryotic translation initiation factor 2, eIF3F, eukaryotic translation initiation factor 3 subunit F |
-0.116 |
-0.011 |
| 90 |
266151_x_at |
AT2G12300
|
[CACTA-like transposase family (Ptta/En/Spm), has a 3.8e-41 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.114 |
0.158 |
| 91 |
264087_at |
AT2G31340
|
emb1381, embryo defective 1381 |
-0.114 |
0.015 |
| 92 |
263020_at |
AT1G23880
|
[NHL domain-containing protein] |
-0.114 |
-0.200 |
| 93 |
246800_at |
AT5G26780
|
SHM2, serine hydroxymethyltransferase 2 |
-0.111 |
-0.188 |
| 94 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
-0.109 |
0.760 |
| 95 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.109 |
-0.054 |
| 96 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.109 |
0.044 |
| 97 |
266744_at |
AT2G02970
|
APY6, apyrase 6, AtAPY6 |
-0.106 |
0.003 |
| 98 |
246770_at |
AT5G27460
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.106 |
-0.073 |
| 99 |
264084_at |
AT2G31240
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.106 |
0.158 |
| 100 |
246788_at |
AT5G27580
|
AGL89, AGAMOUS-like 89 |
-0.103 |
-0.105 |
| 101 |
251202_at |
AT3G63040
|
unknown |
-0.103 |
0.327 |
| 102 |
253021_at |
AT4G38050
|
[Xanthine/uracil permease family protein] |
-0.102 |
0.013 |
| 103 |
260539_at |
AT2G43480
|
[Peroxidase superfamily protein] |
-0.102 |
0.184 |
| 104 |
258128_at |
AT3G24590
|
PLSP1, plastidic type i signal peptidase 1 |
-0.101 |
-0.168 |