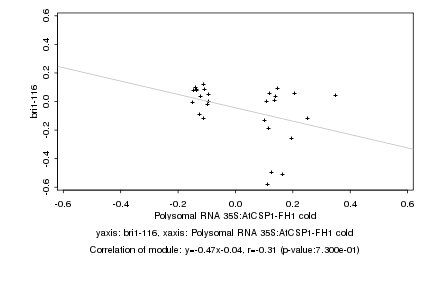
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Polysomal RNA 35S:AtCSP1-FH1 cold |
Log2 signal ratio
bri1-116 |
| 1 |
253129_at |
AT4G36020
|
AtCSP1, CSDP1, cold shock domain protein 1, CSP1, cold shock protein 1 |
0.348 |
0.047 |
| 2 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.250 |
-0.114 |
| 3 |
266751_at |
AT2G47020
|
[Peptide chain release factor 1] |
0.204 |
0.060 |
| 4 |
246827_at |
AT5G26330
|
[Cupredoxin superfamily protein] |
0.192 |
-0.257 |
| 5 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.161 |
-0.507 |
| 6 |
261466_at |
AT1G07690
|
unknown |
0.146 |
0.093 |
| 7 |
257978_at |
AT3G20860
|
ATNEK5, NIMA-related kinase 5, NEK5, NIMA-related kinase 5 |
0.137 |
0.042 |
| 8 |
253690_at |
AT4G29550
|
unknown |
0.134 |
0.013 |
| 9 |
254629_at |
AT4G18425
|
unknown |
0.125 |
-0.494 |
| 10 |
259285_at |
AT3G11460
|
MEF10, mitochondrial RNA editing factor 10 |
0.116 |
0.063 |
| 11 |
257757_at |
AT3G18660
|
GUX1, glucuronic acid substitution of xylan 1, PGSIP1, plant glycogenin-like starch initiation protein 1 |
0.112 |
-0.182 |
| 12 |
251116_at |
AT3G63470
|
scpl40, serine carboxypeptidase-like 40 |
0.110 |
-0.574 |
| 13 |
251568_at |
AT3G58280
|
[Arabidopsis phospholipase-like protein (PEARLI 4) with TRAF-like domain] |
0.108 |
0.003 |
| 14 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
0.100 |
-0.131 |
| 15 |
249533_at |
AT5G38790
|
unknown |
-0.153 |
-0.000 |
| 16 |
267466_at |
AT2G19010
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.147 |
0.082 |
| 17 |
246903_at |
AT5G25750
|
unknown |
-0.141 |
0.103 |
| 18 |
253057_at |
AT4G37670
|
NAGS2, N-acetyl-l-glutamate synthase 2 |
-0.139 |
0.079 |
| 19 |
260013_at |
AT1G68090
|
ANN5, annexin 5, ANNAT5, ANNEXIN ARABIDOPSIS THALIANA 5 |
-0.136 |
0.090 |
| 20 |
265721_at |
AT2G40090
|
ATATH9, ABC2 homolog 9, ATH9, ABC2 homolog 9 |
-0.127 |
-0.085 |
| 21 |
262192_at |
AT1G77830
|
[RING/U-box superfamily protein] |
-0.123 |
0.040 |
| 22 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
-0.113 |
-0.112 |
| 23 |
244907_at |
ATMG00710
|
ORF120 |
-0.112 |
0.120 |
| 24 |
249326_at |
AT5G40840
|
AtRAD21.1, Sister chromatid cohesion 1 (SCC1) protein homolog 2, SYN2 |
-0.109 |
0.091 |
| 25 |
248450_at |
AT5G51290
|
ACD5, ACCELERATED CELL DEATH 5 |
-0.098 |
-0.015 |
| 26 |
249715_at |
AT5G35760
|
[Beta-galactosidase related protein] |
-0.096 |
0.001 |
| 27 |
257450_at |
AT1G10530
|
unknown |
-0.094 |
0.053 |