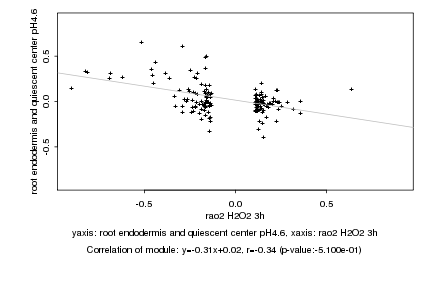
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
rao2 H2O2 3h |
Log2 signal ratio
root endodermis and quiescent center pH4.6 |
| 1 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.633 |
0.142 |
| 2 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
0.357 |
0.011 |
| 3 |
246888_at |
AT5G26270
|
unknown |
0.353 |
-0.128 |
| 4 |
247224_at |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
0.318 |
-0.082 |
| 5 |
262719_at |
AT1G43590
|
unknown |
0.282 |
-0.007 |
| 6 |
265993_at |
AT2G24160
|
[pseudogene] |
0.248 |
-0.045 |
| 7 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
0.240 |
-0.007 |
| 8 |
265893_at |
AT2G15042
|
[Leucine-rich repeat (LRR) family protein] |
0.234 |
-0.003 |
| 9 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
0.232 |
-0.086 |
| 10 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
0.228 |
0.126 |
| 11 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
0.228 |
-0.002 |
| 12 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
0.225 |
-0.212 |
| 13 |
256943_at |
AT3G18910
|
ETP2, EIN2 targeting protein2 |
0.224 |
0.128 |
| 14 |
263674_at |
AT2G04790
|
unknown |
0.217 |
0.005 |
| 15 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
0.204 |
0.043 |
| 16 |
267622_at |
AT2G39690
|
unknown |
0.204 |
0.005 |
| 17 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.201 |
-0.023 |
| 18 |
246947_at |
AT5G25120
|
CYP71B11, ytochrome p450, family 71, subfamily B, polypeptide 11 |
0.190 |
-0.018 |
| 19 |
247223_at |
AT5G65070
|
AGL69, AGAMOUS-like 69, FCL4, MAF4, MADS AFFECTING FLOWERING 4 |
0.184 |
-0.014 |
| 20 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
0.178 |
-0.062 |
| 21 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
0.173 |
-0.032 |
| 22 |
255016_at |
AT4G10120
|
ATSPS4F |
0.168 |
-0.045 |
| 23 |
262750_at |
AT1G28710
|
[Nucleotide-diphospho-sugar transferase family protein] |
0.167 |
-0.169 |
| 24 |
251535_at |
AT3G58540
|
unknown |
0.165 |
0.066 |
| 25 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
0.163 |
0.063 |
| 26 |
266363_at |
AT2G41250
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.156 |
-0.024 |
| 27 |
247977_at |
AT5G56850
|
unknown |
0.153 |
-0.018 |
| 28 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
0.153 |
0.012 |
| 29 |
259753_at |
AT1G71050
|
HIPP20, heavy metal associated isoprenylated plant protein 20 |
0.153 |
-0.388 |
| 30 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
0.151 |
-0.095 |
| 31 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.151 |
-0.025 |
| 32 |
266423_at |
AT2G41340
|
RPB5D, RNA polymerase II fifth largest subunit, D |
0.149 |
-0.013 |
| 33 |
263986_at |
AT2G42790
|
CSY3, citrate synthase 3 |
0.147 |
0.048 |
| 34 |
264343_at |
AT1G11850
|
unknown |
0.145 |
-0.112 |
| 35 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
0.145 |
-0.001 |
| 36 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
0.144 |
-0.234 |
| 37 |
249058_at |
AT5G44510
|
TAO1, target of AVRB operation1 |
0.142 |
0.001 |
| 38 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.140 |
0.103 |
| 39 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
0.140 |
-0.076 |
| 40 |
251759_at |
AT3G55630
|
ATDFD, DHFS-FPGS homolog D, DFD, DHFS-FPGS homolog D, FPGS3, folylpolyglutamate synthetase 3 |
0.138 |
0.206 |
| 41 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
0.138 |
0.082 |
| 42 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
0.138 |
-0.002 |
| 43 |
262786_at |
AT1G10740
|
[alpha/beta-Hydrolases superfamily protein] |
0.136 |
-0.077 |
| 44 |
252838_at |
AT3G42090
|
[contains domain LIN-9 RELATED (PTHR21689)] |
0.135 |
0.030 |
| 45 |
264201_at |
AT1G22630
|
unknown |
0.134 |
-0.046 |
| 46 |
245877_at |
AT1G26220
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.133 |
0.007 |
| 47 |
262396_at |
AT1G49470
|
unknown |
0.131 |
-0.215 |
| 48 |
259901_at |
AT1G74180
|
AtRLP14, receptor like protein 14, RLP14, receptor like protein 14 |
0.129 |
0.069 |
| 49 |
261931_at |
AT1G22430
|
[GroES-like zinc-binding dehydrogenase family protein] |
0.126 |
-0.029 |
| 50 |
260317_at |
AT1G63800
|
UBC5, ubiquitin-conjugating enzyme 5 |
0.126 |
-0.018 |
| 51 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
0.126 |
-0.305 |
| 52 |
250985_at |
AT5G02830
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.125 |
-0.026 |
| 53 |
249212_at |
AT5G42690
|
unknown |
0.123 |
-0.096 |
| 54 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
0.122 |
-0.021 |
| 55 |
262601_at |
AT1G15310
|
ATHSRP54A, signal recognition particle 54 kDa subunit, SRP54-1, SIGNAL RECOGNITION PARTICLE 54 KDA SUBUNIT |
0.121 |
0.020 |
| 56 |
257174_at |
AT3G27190
|
UKL2, uridine kinase-like 2 |
0.121 |
-0.056 |
| 57 |
254870_at |
AT4G11900
|
[S-locus lectin protein kinase family protein] |
0.121 |
-0.031 |
| 58 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
0.121 |
-0.104 |
| 59 |
260086_at |
AT1G63240
|
unknown |
0.120 |
-0.030 |
| 60 |
264096_at |
AT1G78995
|
unknown |
0.119 |
-0.008 |
| 61 |
259956_at |
AT1G75110
|
RRA2, REDUCED RESIDUAL ARABINOSE 2 |
0.118 |
-0.104 |
| 62 |
263264_at |
AT2G38810
|
HTA8, histone H2A 8 |
0.117 |
0.004 |
| 63 |
253909_at |
AT4G27270
|
[Quinone reductase family protein] |
0.116 |
-0.080 |
| 64 |
254817_at |
AT4G12460
|
ORP2B, OSBP(oxysterol binding protein)-related protein 2B |
0.116 |
-0.054 |
| 65 |
251741_at |
AT3G56040
|
UGP3, UDP-glucose pyrophosphorylase 3 |
0.116 |
0.032 |
| 66 |
261056_at |
AT1G01360
|
PYL9, PYRABACTIN RESISTANCE 1-LIKE 9, RCAR1, regulatory component of ABA receptor 1 |
0.115 |
-0.028 |
| 67 |
254479_at |
AT4G20350
|
[oxidoreductases] |
0.115 |
0.080 |
| 68 |
249639_at |
AT5G36930
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.114 |
0.068 |
| 69 |
257966_at |
AT3G19800
|
unknown |
0.114 |
-0.064 |
| 70 |
264405_at |
AT1G10210
|
ATMPK1, mitogen-activated protein kinase 1, MPK1, mitogen-activated protein kinase 1 |
0.112 |
-0.106 |
| 71 |
259902_at |
AT1G74170
|
AtRLP13, receptor like protein 13, RLP13, receptor like protein 13 |
0.112 |
0.001 |
| 72 |
264435_at |
AT1G10360
|
ATGSTU18, glutathione S-transferase TAU 18, GST29, GLUTATHIONE S-TRANSFERASE 29, GSTU18, glutathione S-transferase TAU 18 |
0.112 |
-0.092 |
| 73 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
0.112 |
0.004 |
| 74 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.111 |
0.043 |
| 75 |
258326_at |
AT3G22760
|
SOL1 |
0.110 |
-0.100 |
| 76 |
259907_at |
AT1G60860
|
AGD2, ARF-GAP domain 2 |
0.108 |
0.076 |
| 77 |
261888_at |
AT1G80800
|
[pseudogene] |
0.107 |
0.042 |
| 78 |
265720_at |
AT2G40110
|
[Yippee family putative zinc-binding protein] |
0.107 |
-0.091 |
| 79 |
260364_at |
AT1G70560
|
CKRC1, cytokinin induced root curling 1, SAV3, SHADE AVOIDANCE 3, TAA1, tryptophan aminotransferase of Arabidopsis 1, WEI8, WEAK ETHYLENE INSENSITIVE 8 |
0.107 |
-0.023 |
| 80 |
266959_at |
AT2G07690
|
MCM5, MINICHROMOSOME MAINTENANCE 5 |
0.105 |
-0.005 |
| 81 |
257745_at |
AT3G29240
|
unknown |
0.105 |
0.138 |
| 82 |
249542_at |
AT5G38140
|
NF-YC12, nuclear factor Y, subunit C12 |
0.105 |
-0.002 |
| 83 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
-0.904 |
0.145 |
| 84 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
-0.824 |
0.337 |
| 85 |
260522_x_at |
AT2G41730
|
unknown |
-0.813 |
0.330 |
| 86 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
-0.696 |
0.261 |
| 87 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.686 |
0.313 |
| 88 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
-0.625 |
0.273 |
| 89 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.519 |
0.654 |
| 90 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
-0.461 |
0.363 |
| 91 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
-0.456 |
0.293 |
| 92 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
-0.450 |
0.207 |
| 93 |
263515_at |
AT2G21640
|
unknown |
-0.443 |
0.439 |
| 94 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
-0.386 |
0.312 |
| 95 |
266618_at |
AT2G35480
|
unknown |
-0.365 |
0.258 |
| 96 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
-0.340 |
0.061 |
| 97 |
245999_at |
AT5G20650
|
COPT5, copper transporter 5 |
-0.330 |
-0.047 |
| 98 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
-0.309 |
0.128 |
| 99 |
256442_at |
AT3G10930
|
unknown |
-0.295 |
-0.044 |
| 100 |
251513_at |
AT3G59220
|
ATPIRIN1, PRN, pirin, PRN1 |
-0.293 |
-0.116 |
| 101 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.291 |
0.611 |
| 102 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
-0.283 |
0.030 |
| 103 |
254010_at |
AT4G26240
|
unknown |
-0.274 |
0.009 |
| 104 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.265 |
0.033 |
| 105 |
248253_at |
AT5G53290
|
CRF3, cytokinin response factor 3 |
-0.263 |
0.143 |
| 106 |
257401_at |
AT1G23550
|
SRO2, similar to RCD one 2 |
-0.254 |
0.120 |
| 107 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
-0.248 |
0.349 |
| 108 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
-0.244 |
-0.119 |
| 109 |
254200_at |
AT4G24110
|
unknown |
-0.241 |
0.018 |
| 110 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
-0.238 |
-0.063 |
| 111 |
254603_at |
AT4G19045
|
[Mob1/phocein family protein] |
-0.231 |
-0.109 |
| 112 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
-0.231 |
0.101 |
| 113 |
264460_at |
AT1G10170
|
ATNFXL1, NF-X-like 1, NFXL1, NF-X-like 1 |
-0.225 |
0.269 |
| 114 |
244901_at |
ATMG00640
|
ORF25 |
-0.223 |
0.092 |
| 115 |
250479_at |
AT5G10260
|
AtRABH1e, RAB GTPase homolog H1E, RABH1e, RAB GTPase homolog H1E |
-0.222 |
-0.062 |
| 116 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
-0.222 |
-0.053 |
| 117 |
267140_at |
AT2G38250
|
[Homeodomain-like superfamily protein] |
-0.217 |
0.261 |
| 118 |
262514_at |
AT1G34190
|
anac017, NAC domain containing protein 17, NAC017, NAC domain containing protein 17 |
-0.215 |
-0.001 |
| 119 |
258397_at |
AT3G15357
|
unknown |
-0.213 |
0.314 |
| 120 |
247287_at |
AT5G64230
|
unknown |
-0.212 |
0.081 |
| 121 |
266606_at |
AT2G46310
|
CRF5, cytokinin response factor 5 |
-0.199 |
-0.030 |
| 122 |
250907_at |
AT5G03670
|
TRM28, TON1 Recruiting Motif 28 |
-0.198 |
-0.128 |
| 123 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
-0.190 |
-0.193 |
| 124 |
261187_at |
AT1G32860
|
[Glycosyl hydrolase superfamily protein] |
-0.190 |
0.002 |
| 125 |
256725_at |
AT2G34070
|
TBL37, TRICHOME BIREFRINGENCE-LIKE 37 |
-0.188 |
-0.085 |
| 126 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
-0.187 |
0.196 |
| 127 |
263061_at |
AT2G18190
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.182 |
-0.024 |
| 128 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.175 |
-0.091 |
| 129 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
-0.175 |
0.115 |
| 130 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
-0.172 |
-0.033 |
| 131 |
247447_at |
AT5G62730
|
[Major facilitator superfamily protein] |
-0.170 |
-0.051 |
| 132 |
246282_at |
AT4G36580
|
[AAA-type ATPase family protein] |
-0.169 |
0.495 |
| 133 |
250194_at |
AT5G14550
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.168 |
-0.021 |
| 134 |
249068_at |
AT5G43980
|
PDLP1, plasmodesmata-located protein 1, PDLP1A, PLASMODESMATA-LOCATED PROTEIN 1A |
-0.168 |
-0.008 |
| 135 |
258805_at |
AT3G04010
|
[O-Glycosyl hydrolases family 17 protein] |
-0.168 |
-0.049 |
| 136 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
-0.168 |
-0.144 |
| 137 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
-0.165 |
-0.060 |
| 138 |
253824_at |
AT4G27940
|
ATMTM1, ARABIDOPSIS MANGANESE TRACKING FACTOR FOR MITOCHONDRIAL SOD2, MTM1, manganese tracking factor for mitochondrial SOD2 |
-0.165 |
0.186 |
| 139 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
-0.165 |
0.374 |
| 140 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
-0.165 |
0.098 |
| 141 |
251603_at |
AT3G57760
|
[Protein kinase superfamily protein] |
-0.164 |
0.055 |
| 142 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
-0.163 |
0.501 |
| 143 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-0.161 |
0.019 |
| 144 |
263614_at |
AT2G25240
|
AtCCP3, CCP3, conserved in ciliated species and in the land plants 3 |
-0.159 |
0.060 |
| 145 |
260415_at |
AT1G69790
|
[Protein kinase superfamily protein] |
-0.159 |
0.138 |
| 146 |
259222_at |
AT3G03680
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.156 |
0.068 |
| 147 |
262836_at |
AT1G14680
|
unknown |
-0.155 |
0.052 |
| 148 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-0.155 |
-0.005 |
| 149 |
259626_at |
AT1G42990
|
ATBZIP60, basic region/leucine zipper motif 60, BZIP60, basic region/leucine zipper motif 60, BZIP60, Bbasic region/leucine zipper motif 60 |
-0.150 |
-0.003 |
| 150 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
-0.150 |
-0.114 |
| 151 |
263822_at |
AT2G40240
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.148 |
0.107 |
| 152 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
-0.148 |
-0.009 |
| 153 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
-0.147 |
-0.038 |
| 154 |
250821_at |
AT5G05190
|
unknown |
-0.147 |
-0.049 |
| 155 |
261261_at |
AT1G26730
|
[EXS (ERD1/XPR1/SYG1) family protein] |
-0.146 |
-0.187 |
| 156 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.146 |
-0.326 |
| 157 |
248101_at |
AT5G55200
|
MGE1, mitochondrial GrpE 1 |
-0.145 |
0.182 |
| 158 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.142 |
-0.017 |
| 159 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
-0.141 |
-0.171 |
| 160 |
253405_at |
AT4G32800
|
[Integrase-type DNA-binding superfamily protein] |
-0.140 |
0.085 |
| 161 |
255811_at |
AT4G10250
|
ATHSP22.0 |
-0.139 |
-0.213 |
| 162 |
247933_at |
AT5G56980
|
unknown |
-0.139 |
0.052 |
| 163 |
247989_at |
AT5G56350
|
[Pyruvate kinase family protein] |
-0.132 |
0.098 |
| 164 |
261063_at |
AT1G07520
|
[GRAS family transcription factor] |
-0.132 |
-0.037 |