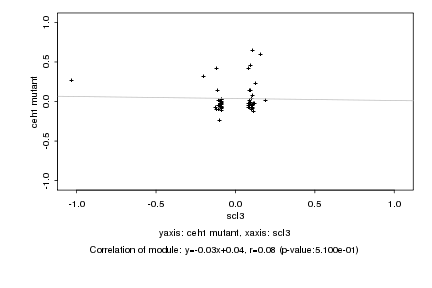
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
scl3 |
Log2 signal ratio
ceh1 mutant |
| 1 |
265332_at |
AT2G18410
|
unknown |
0.186 |
0.025 |
| 2 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.154 |
0.597 |
| 3 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.126 |
0.240 |
| 4 |
267215_at |
AT2G02630
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.115 |
-0.023 |
| 5 |
256588_at |
AT3G28790
|
unknown |
0.110 |
-0.126 |
| 6 |
244994_at |
ATCG01010
|
NDHF |
0.108 |
-0.021 |
| 7 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.107 |
0.084 |
| 8 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.107 |
0.653 |
| 9 |
262332_at |
AT1G64030
|
ATSRP3, SRP3, serpin 3 |
0.104 |
-0.031 |
| 10 |
252614_at |
AT3G45220
|
[Serine protease inhibitor (SERPIN) family protein] |
0.104 |
-0.065 |
| 11 |
265367_at |
AT2G13270
|
unknown |
0.104 |
-0.085 |
| 12 |
262740_at |
AT1G28590
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.103 |
-0.050 |
| 13 |
245579_at |
AT4G14810
|
unknown |
0.101 |
-0.040 |
| 14 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.098 |
0.049 |
| 15 |
245133_at |
AT2G45310
|
GAE4, UDP-D-glucuronate 4-epimerase 4 |
0.096 |
-0.100 |
| 16 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.093 |
0.143 |
| 17 |
249413_at |
AT5G39620
|
AtRABG1, RAB GTPase homolog G1, RABG1, RAB GTPase homolog G1 |
0.092 |
-0.027 |
| 18 |
249714_at |
AT5G35710
|
[copia-like retrotransposon family, has a 2.5e-48 P-value blast match to dbj|BAA78425.1| polyprotein (Arabidopsis thaliana) (AtRE1) (Ty1_Copia-element)] |
0.092 |
-0.006 |
| 19 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
0.090 |
0.457 |
| 20 |
254677_at |
AT4G18490
|
unknown |
0.087 |
-0.044 |
| 21 |
262653_at |
AT1G14130
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.085 |
0.142 |
| 22 |
265625_at |
AT2G27240
|
[Aluminium activated malate transporter family protein] |
0.084 |
-0.041 |
| 23 |
248660_at |
AT5G48650
|
[Nuclear transport factor 2 (NTF2) family protein with RNA binding (RRM-RBD-RNP motifs) domain] |
0.084 |
0.023 |
| 24 |
252517_at |
AT3G46340
|
[Leucine-rich repeat protein kinase family protein] |
0.084 |
-0.066 |
| 25 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.082 |
0.428 |
| 26 |
263266_at |
AT2G16520
|
unknown |
0.081 |
-0.048 |
| 27 |
261198_at |
AT1G12940
|
ATNRT2.5, nitrate transporter2.5, NRT2.5, nitrate transporter2.5 |
0.080 |
-0.066 |
| 28 |
258363_at |
AT3G14300
|
ATPME26, A. THALIANA PECTIN METHYLESTERASE 26, ATPMEPCRC, PME26, PECTIN METHYLESTERASE 26 |
0.076 |
-0.013 |
| 29 |
261866_at |
AT1G50420
|
SCL-3, SCARECROW-LIKE 3, SCL3, scarecrow-like 3 |
-1.037 |
0.271 |
| 30 |
253832_at |
AT4G27654
|
unknown |
-0.204 |
0.319 |
| 31 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
-0.131 |
-0.070 |
| 32 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
-0.123 |
0.430 |
| 33 |
256659_at |
AT3G12020
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.120 |
-0.094 |
| 34 |
264251_at |
AT1G09190
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.116 |
0.144 |
| 35 |
267626_at |
AT2G42250
|
CYP712A1, cytochrome P450, family 712, subfamily A, polypeptide 1 |
-0.112 |
-0.038 |
| 36 |
248008_at |
AT5G56270
|
ATWRKY2, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 2, WRKY2, WRKY DNA-binding protein 2 |
-0.110 |
0.015 |
| 37 |
250810_at |
AT5G05090
|
[Homeodomain-like superfamily protein] |
-0.109 |
-0.100 |
| 38 |
251613_at |
AT3G57960
|
[Emsy N Terminus (ENT) domain-containing protein] |
-0.107 |
-0.049 |
| 39 |
255957_at |
AT1G22160
|
unknown |
-0.104 |
-0.229 |
| 40 |
265094_at |
AT1G03890
|
[RmlC-like cupins superfamily protein] |
-0.100 |
-0.013 |
| 41 |
263959_at |
AT2G36210
|
SAUR45, SMALL AUXIN UPREGULATED RNA 45 |
-0.097 |
0.012 |
| 42 |
260218_at |
AT1G74620
|
[RING/U-box superfamily protein] |
-0.097 |
-0.058 |
| 43 |
266144_at |
AT2G12170
|
unknown |
-0.095 |
-0.022 |
| 44 |
261246_at |
AT1G20080
|
ATSYTB, NTMC2T1.2, NTMC2TYPE1.2, SYT2, synaptotagmin 2, SYTB |
-0.095 |
-0.031 |
| 45 |
246866_at |
AT5G25960
|
unknown |
-0.094 |
-0.013 |
| 46 |
254441_at |
AT4G21050
|
AtDOF4.4, DOF4.4, DNA-binding with One Finger 4.4 |
-0.094 |
0.012 |
| 47 |
255765_at |
AT1G16760
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.093 |
-0.058 |
| 48 |
247501_at |
AT5G61920
|
FLL4, FLOWERING LOCUS C EXPRESSOR-LIKE 4, FLX4, FLX LIKE 4 |
-0.093 |
0.030 |
| 49 |
248982_at |
AT5G45120
|
[Eukaryotic aspartyl protease family protein] |
-0.093 |
-0.064 |
| 50 |
249470_at |
AT5G39350
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.091 |
0.037 |
| 51 |
250331_at |
AT5G11820
|
[Plant self-incompatibility protein S1 family] |
-0.091 |
-0.012 |
| 52 |
254482_at |
AT4G20370
|
TSF, TWIN SISTER OF FT |
-0.090 |
-0.103 |
| 53 |
246706_at |
AT5G28130
|
unknown |
-0.090 |
-0.036 |
| 54 |
247613_at |
AT5G60740
|
ABCG28, ATP-binding cassette G28 |
-0.089 |
-0.083 |
| 55 |
262261_at |
AT1G70895
|
CLE17, CLAVATA3/ESR-RELATED 17 |
-0.089 |
-0.068 |