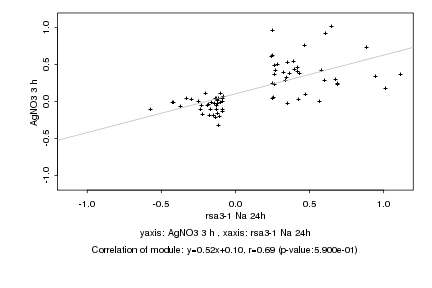
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
rsa3-1 Na 24h |
Log2 signal ratio
AgNO3 3 h |
| 1 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
1.111 |
0.373 |
| 2 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
1.011 |
0.179 |
| 3 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.944 |
0.347 |
| 4 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.883 |
0.735 |
| 5 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.688 |
0.246 |
| 6 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.684 |
0.235 |
| 7 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.675 |
0.309 |
| 8 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
0.647 |
1.028 |
| 9 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.601 |
0.935 |
| 10 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
0.597 |
0.297 |
| 11 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.575 |
0.423 |
| 12 |
265611_at |
AT2G25510
|
unknown |
0.566 |
0.002 |
| 13 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.470 |
0.108 |
| 14 |
262085_at |
AT1G56060
|
unknown |
0.461 |
0.765 |
| 15 |
266017_at |
AT2G18690
|
unknown |
0.426 |
0.381 |
| 16 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.423 |
0.040 |
| 17 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.413 |
0.416 |
| 18 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
0.413 |
0.470 |
| 19 |
253632_at |
AT4G30430
|
TET9, tetraspanin9 |
0.393 |
0.434 |
| 20 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.389 |
0.549 |
| 21 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.363 |
0.384 |
| 22 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
0.347 |
-0.022 |
| 23 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.345 |
0.531 |
| 24 |
260744_at |
AT1G15010
|
unknown |
0.340 |
0.338 |
| 25 |
264635_at |
AT1G65500
|
unknown |
0.337 |
0.290 |
| 26 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.321 |
0.397 |
| 27 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.282 |
0.505 |
| 28 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.268 |
0.427 |
| 29 |
260419_at |
AT1G69730
|
[Wall-associated kinase family protein] |
0.263 |
0.234 |
| 30 |
246018_at |
AT5G10695
|
unknown |
0.263 |
0.375 |
| 31 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.257 |
0.495 |
| 32 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.251 |
0.064 |
| 33 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
0.250 |
0.248 |
| 34 |
259173_at |
AT3G03640
|
BGLU25, beta glucosidase 25, GLUC |
0.249 |
0.041 |
| 35 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.248 |
0.969 |
| 36 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.247 |
0.630 |
| 37 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.243 |
0.614 |
| 38 |
257130_at |
AT3G20210
|
DELTA-VPE, delta vacuolar processing enzyme, DELTAVPE, DELTA VACUOLAR PROCESSING ENZYME |
-0.576 |
-0.096 |
| 39 |
265891_at |
AT2G15010
|
[Plant thionin] |
-0.426 |
-0.011 |
| 40 |
256139_at |
AT1G48660
|
[Auxin-responsive GH3 family protein] |
-0.423 |
-0.003 |
| 41 |
265283_at |
AT2G20370
|
AtMUR3, KAM1, KATAMARI 1, MUR3, MURUS 3, RSA3, SHORT ROOT IN SALT MEDIUM 3 |
-0.371 |
-0.062 |
| 42 |
248435_at |
AT5G51210
|
OLEO3, oleosin3 |
-0.335 |
0.050 |
| 43 |
249937_at |
AT5G22470
|
[NAD+ ADP-ribosyltransferases] |
-0.303 |
0.031 |
| 44 |
263881_at |
AT2G21820
|
unknown |
-0.250 |
0.010 |
| 45 |
259574_at |
AT1G35310
|
MLP168, MLP-like protein 168 |
-0.238 |
-0.108 |
| 46 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.233 |
-0.052 |
| 47 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.229 |
-0.173 |
| 48 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
-0.207 |
0.112 |
| 49 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.195 |
-0.046 |
| 50 |
260088_at |
AT1G73190
|
ALPHA-TIP, ALPHA-TONOPLAST INTRINSIC PROTEIN, TIP3;1 |
-0.187 |
-0.038 |
| 51 |
258013_at |
AT3G19320
|
[Leucine-rich repeat (LRR) family protein] |
-0.180 |
-0.177 |
| 52 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
-0.171 |
-0.105 |
| 53 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.165 |
-0.011 |
| 54 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.153 |
-0.178 |
| 55 |
254677_at |
AT4G18490
|
unknown |
-0.142 |
-0.015 |
| 56 |
257197_at |
AT3G23800
|
SBP3, selenium-binding protein 3 |
-0.140 |
-0.212 |
| 57 |
259615_at |
AT1G47980
|
unknown |
-0.135 |
0.050 |
| 58 |
263544_at |
AT2G21590
|
APL4 |
-0.131 |
-0.044 |
| 59 |
245233_at |
AT4G25580
|
[CAP160 protein] |
-0.131 |
0.054 |
| 60 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.130 |
-0.096 |
| 61 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
-0.127 |
0.027 |
| 62 |
263573_at |
AT2G17150
|
NLP1, NIN-like protein 1 |
-0.125 |
-0.158 |
| 63 |
244997_at |
ATCG00170
|
RPOC2 |
-0.123 |
-0.016 |
| 64 |
266191_at |
AT2G39040
|
[Peroxidase superfamily protein] |
-0.120 |
-0.312 |
| 65 |
245232_at |
AT4G25590
|
ADF7, actin depolymerizing factor 7 |
-0.115 |
0.043 |
| 66 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.112 |
-0.193 |
| 67 |
255950_at |
AT1G22110
|
[structural constituent of ribosome] |
-0.106 |
0.110 |
| 68 |
261616_at |
AT1G33060
|
ANAC014, NAC 014, NAC014, NAC 014 |
-0.106 |
-0.007 |
| 69 |
252705_at |
AT3G43760
|
unknown |
-0.094 |
0.049 |
| 70 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
-0.093 |
-0.135 |
| 71 |
259961_at |
AT1G53700
|
PK3AT, PROTEIN KINASE 3 ARABIDOPSIS THALIANA, WAG1, WAG 1 |
-0.089 |
0.004 |
| 72 |
254588_at |
AT4G18840
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.089 |
-0.107 |
| 73 |
260752_at |
AT1G49030
|
[PLAC8 family protein] |
-0.086 |
0.078 |