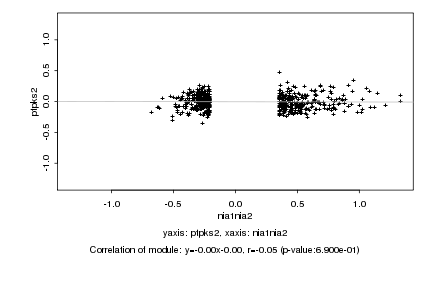
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
nia1nia2 |
Log2 signal ratio
ptpks2 |
| 1 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
1.329 |
0.006 |
| 2 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
1.325 |
0.112 |
| 3 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
1.211 |
-0.051 |
| 4 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
1.140 |
0.143 |
| 5 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
1.120 |
-0.094 |
| 6 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
1.086 |
-0.085 |
| 7 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
1.076 |
0.177 |
| 8 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
1.057 |
0.213 |
| 9 |
257481_at |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
1.024 |
-0.117 |
| 10 |
257517_at |
AT3G16330
|
unknown |
1.019 |
0.040 |
| 11 |
245799_at |
AT1G45616
|
AtRLP6, receptor like protein 6, RLP6, receptor like protein 6 |
1.010 |
-0.172 |
| 12 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.996 |
-0.062 |
| 13 |
265922_at |
AT2G18480
|
[Major facilitator superfamily protein] |
0.979 |
-0.166 |
| 14 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
0.948 |
0.349 |
| 15 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.939 |
0.171 |
| 16 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.935 |
-0.038 |
| 17 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.912 |
0.265 |
| 18 |
245912_at |
AT5G19600
|
SULTR3;5, sulfate transporter 3;5 |
0.910 |
-0.069 |
| 19 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.909 |
-0.069 |
| 20 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.882 |
0.038 |
| 21 |
254743_at |
AT4G13420
|
ATHAK5, ARABIDOPSIS THALIANA HIGH AFFINITY K+ TRANSPORTER 5, HAK5, high affinity K+ transporter 5 |
0.879 |
-0.148 |
| 22 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.878 |
-0.022 |
| 23 |
262373_at |
AT1G73120
|
unknown |
0.873 |
-0.028 |
| 24 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
0.871 |
0.098 |
| 25 |
254634_at |
AT4G18650
|
[transcription factor-related] |
0.859 |
0.039 |
| 26 |
249454_at |
AT5G39520
|
unknown |
0.857 |
0.030 |
| 27 |
263178_at |
AT1G05550
|
unknown |
0.838 |
0.060 |
| 28 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.833 |
-0.030 |
| 29 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.826 |
-0.038 |
| 30 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.812 |
0.033 |
| 31 |
247047_at |
AT5G66650
|
unknown |
0.807 |
-0.126 |
| 32 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.799 |
-0.111 |
| 33 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.797 |
0.006 |
| 34 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.793 |
0.005 |
| 35 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.791 |
-0.107 |
| 36 |
247035_at |
AT5G67110
|
ALC, ALCATRAZ |
0.787 |
-0.045 |
| 37 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.785 |
-0.204 |
| 38 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.785 |
0.242 |
| 39 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
0.784 |
-0.083 |
| 40 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.774 |
0.035 |
| 41 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.773 |
0.138 |
| 42 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.772 |
-0.143 |
| 43 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.765 |
0.251 |
| 44 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.763 |
0.178 |
| 45 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
0.759 |
-0.101 |
| 46 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.759 |
-0.100 |
| 47 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.752 |
-0.051 |
| 48 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.748 |
0.081 |
| 49 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.735 |
-0.116 |
| 50 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.715 |
-0.047 |
| 51 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.713 |
-0.062 |
| 52 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.713 |
-0.097 |
| 53 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.704 |
0.185 |
| 54 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.703 |
-0.011 |
| 55 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.691 |
0.167 |
| 56 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.680 |
0.263 |
| 57 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.679 |
0.254 |
| 58 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.679 |
-0.036 |
| 59 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.675 |
-0.115 |
| 60 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.671 |
-0.006 |
| 61 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.664 |
-0.002 |
| 62 |
263061_at |
AT2G18190
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.659 |
-0.011 |
| 63 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.649 |
0.106 |
| 64 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
0.647 |
-0.114 |
| 65 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.643 |
-0.125 |
| 66 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.642 |
0.183 |
| 67 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.641 |
0.110 |
| 68 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.640 |
-0.002 |
| 69 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.639 |
0.118 |
| 70 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.635 |
0.175 |
| 71 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
0.634 |
-0.191 |
| 72 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.634 |
0.028 |
| 73 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.633 |
0.034 |
| 74 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.633 |
0.033 |
| 75 |
256725_at |
AT2G34070
|
TBL37, TRICHOME BIREFRINGENCE-LIKE 37 |
0.631 |
0.013 |
| 76 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.617 |
-0.003 |
| 77 |
253047_at |
AT4G37295
|
unknown |
0.614 |
-0.069 |
| 78 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.613 |
0.180 |
| 79 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.613 |
-0.084 |
| 80 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.610 |
-0.021 |
| 81 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.590 |
-0.132 |
| 82 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.587 |
-0.001 |
| 83 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.584 |
-0.123 |
| 84 |
264720_at |
AT1G70080
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.580 |
-0.245 |
| 85 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.580 |
0.109 |
| 86 |
252539_at |
AT3G45730
|
unknown |
0.575 |
-0.025 |
| 87 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.574 |
-0.002 |
| 88 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.573 |
-0.115 |
| 89 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
0.573 |
0.087 |
| 90 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.572 |
-0.108 |
| 91 |
260522_x_at |
AT2G41730
|
unknown |
0.571 |
-0.196 |
| 92 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.565 |
0.049 |
| 93 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.564 |
0.104 |
| 94 |
251971_at |
AT3G53160
|
UGT73C7, UDP-glucosyl transferase 73C7 |
0.559 |
-0.127 |
| 95 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.558 |
-0.189 |
| 96 |
263515_at |
AT2G21640
|
unknown |
0.557 |
0.103 |
| 97 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.553 |
0.248 |
| 98 |
264288_at |
AT1G62045
|
unknown |
0.553 |
-0.037 |
| 99 |
265151_at |
AT1G51340
|
[MATE efflux family protein] |
0.549 |
0.060 |
| 100 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
0.539 |
0.107 |
| 101 |
261585_at |
AT1G01010
|
ANAC001, NAC domain containing protein 1, NAC001, NAC domain containing protein 1 |
0.538 |
-0.004 |
| 102 |
264186_at |
AT1G54570
|
PES1, phytyl ester synthase 1 |
0.537 |
0.074 |
| 103 |
246374_at |
AT1G51840
|
[protein kinase-related] |
0.536 |
-0.049 |
| 104 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
0.535 |
-0.006 |
| 105 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
0.534 |
-0.073 |
| 106 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.531 |
0.076 |
| 107 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.530 |
-0.125 |
| 108 |
244939_at |
ATCG00065
|
RPS12, RIBOSOMAL PROTEIN S12, RPS12A, ribosomal protein S12A |
0.529 |
-0.193 |
| 109 |
251513_at |
AT3G59220
|
ATPIRIN1, PRN, pirin, PRN1 |
0.529 |
-0.067 |
| 110 |
264635_at |
AT1G65500
|
unknown |
0.528 |
-0.092 |
| 111 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.527 |
-0.023 |
| 112 |
254630_at |
AT4G18360
|
GOX3, glycolate oxidase 3 |
0.525 |
-0.105 |
| 113 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.521 |
-0.153 |
| 114 |
247951_at |
AT5G57240
|
ORP4C, OSBP(oxysterol binding protein)-related protein 4C |
0.521 |
-0.192 |
| 115 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.520 |
-0.164 |
| 116 |
265999_at |
AT2G24100
|
ASG1, ALTERED SEED GERMINATION 1 |
0.518 |
-0.064 |
| 117 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
0.516 |
-0.068 |
| 118 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.515 |
0.118 |
| 119 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.513 |
-0.033 |
| 120 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.509 |
-0.057 |
| 121 |
265025_at |
AT1G24575
|
unknown |
0.508 |
0.139 |
| 122 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.507 |
0.048 |
| 123 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.504 |
0.037 |
| 124 |
264245_at |
AT1G60450
|
AtGolS7, galactinol synthase 7, GolS7, galactinol synthase 7 |
0.504 |
-0.135 |
| 125 |
267266_at |
AT2G23150
|
ATNRAMP3, NRAMP3, natural resistance-associated macrophage protein 3 |
0.503 |
-0.070 |
| 126 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.502 |
-0.091 |
| 127 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.499 |
-0.010 |
| 128 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.493 |
-0.183 |
| 129 |
255230_at |
AT4G05390
|
ATRFNR1, root FNR 1, RFNR1, root FNR 1 |
0.491 |
0.072 |
| 130 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.491 |
-0.064 |
| 131 |
245903_at |
AT5G11100
|
ATSYTD, NTMC2T2.2, NTMC2TYPE2.2, SYT4, synaptotagmin 4, SYTD |
0.491 |
-0.033 |
| 132 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.488 |
-0.030 |
| 133 |
247519_at |
AT5G61430
|
ANAC100, NAC domain containing protein 100, ATNAC5, NAC100, NAC domain containing protein 100 |
0.487 |
-0.091 |
| 134 |
250267_at |
AT5G12930
|
unknown |
0.485 |
-0.067 |
| 135 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.483 |
-0.144 |
| 136 |
251503_at |
AT3G59140
|
ABCC10, ATP-binding cassette C10, ATMRP14, multidrug resistance-associated protein 14, MRP14, multidrug resistance-associated protein 14 |
0.482 |
0.086 |
| 137 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.480 |
-0.138 |
| 138 |
248253_at |
AT5G53290
|
CRF3, cytokinin response factor 3 |
0.479 |
-0.102 |
| 139 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
0.477 |
-0.162 |
| 140 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.477 |
0.238 |
| 141 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.471 |
-0.186 |
| 142 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
0.470 |
-0.006 |
| 143 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
0.470 |
0.138 |
| 144 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
0.469 |
-0.182 |
| 145 |
266464_at |
AT2G47800
|
ABCC4, ATP-binding cassette C4, ATMRP4, multidrug resistance-associated protein 4, EST3, MRP4, multidrug resistance-associated protein 4 |
0.467 |
0.245 |
| 146 |
256525_at |
AT1G66180
|
[Eukaryotic aspartyl protease family protein] |
0.463 |
0.063 |
| 147 |
255032_at |
AT4G09500
|
[UDP-Glycosyltransferase superfamily protein] |
0.463 |
-0.210 |
| 148 |
258682_at |
AT3G08720
|
ATPK19, Arabidopsis thaliana protein kinase 19, ATPK2, ARABIDOPSIS THALIANA PROTEIN KINASE 2, ATS6K2, ARABIDOPSIS THALIANA SERINE/THREONINE PROTEIN KINASE 2, S6K2, serine/threonine protein kinase 2 |
0.460 |
0.037 |
| 149 |
265276_at |
AT2G28400
|
unknown |
0.459 |
-0.034 |
| 150 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.458 |
-0.166 |
| 151 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.457 |
-0.045 |
| 152 |
246125_at |
AT5G19875
|
unknown |
0.457 |
0.083 |
| 153 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.456 |
-0.052 |
| 154 |
246495_at |
AT5G16200
|
[50S ribosomal protein-related] |
0.456 |
-0.001 |
| 155 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.453 |
-0.097 |
| 156 |
256296_at |
AT1G69480
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.452 |
-0.138 |
| 157 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.452 |
0.037 |
| 158 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.452 |
-0.035 |
| 159 |
259911_at |
AT1G72680
|
ATCAD1, CINNAMYL ALCOHOL DEHYDROGENASE 1, CAD1, cinnamyl-alcohol dehydrogenase |
0.451 |
0.194 |
| 160 |
261946_at |
AT1G64560
|
|
0.449 |
-0.115 |
| 161 |
254317_at |
AT4G22510
|
unknown |
0.446 |
0.036 |
| 162 |
252882_at |
AT4G39675
|
unknown |
0.446 |
-0.182 |
| 163 |
252316_at |
AT3G48700
|
ATCXE13, carboxyesterase 13, CXE13, carboxyesterase 13 |
0.444 |
-0.009 |
| 164 |
257331_at |
ATMG01330
|
ORF107H |
0.444 |
0.069 |
| 165 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.442 |
0.025 |
| 166 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.440 |
-0.071 |
| 167 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
0.439 |
-0.075 |
| 168 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.438 |
-0.170 |
| 169 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.438 |
-0.090 |
| 170 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.434 |
0.131 |
| 171 |
264580_at |
AT1G05340
|
unknown |
0.434 |
-0.151 |
| 172 |
249068_at |
AT5G43980
|
PDLP1, plasmodesmata-located protein 1, PDLP1A, PLASMODESMATA-LOCATED PROTEIN 1A |
0.433 |
0.050 |
| 173 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.432 |
0.106 |
| 174 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.432 |
0.172 |
| 175 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.430 |
0.033 |
| 176 |
262137_at |
AT1G77920
|
TGA7, TGACG sequence-specific binding protein 7 |
0.429 |
0.020 |
| 177 |
261110_at |
AT1G75440
|
UBC16, ubiquitin-conjugating enzyme 16 |
0.427 |
0.041 |
| 178 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
0.427 |
-0.196 |
| 179 |
253638_at |
AT4G30470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.424 |
0.067 |
| 180 |
264752_at |
AT1G23010
|
LPR1, Low Phosphate Root1 |
0.422 |
-0.074 |
| 181 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.422 |
0.004 |
| 182 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.421 |
0.221 |
| 183 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.419 |
0.310 |
| 184 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.417 |
0.041 |
| 185 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
0.414 |
0.163 |
| 186 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.413 |
0.166 |
| 187 |
266606_at |
AT2G46310
|
CRF5, cytokinin response factor 5 |
0.412 |
-0.075 |
| 188 |
248614_at |
AT5G49560
|
[Putative methyltransferase family protein] |
0.411 |
-0.117 |
| 189 |
250252_at |
AT5G13750
|
ZIFL1, zinc induced facilitator-like 1 |
0.410 |
-0.058 |
| 190 |
252859_at |
AT4G39780
|
[Integrase-type DNA-binding superfamily protein] |
0.406 |
0.100 |
| 191 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.405 |
-0.017 |
| 192 |
260005_at |
AT1G67920
|
unknown |
0.405 |
-0.080 |
| 193 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.405 |
-0.058 |
| 194 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
0.405 |
-0.073 |
| 195 |
253582_at |
AT4G30670
|
[Putative membrane lipoprotein] |
0.405 |
-0.231 |
| 196 |
245882_at |
AT5G09470
|
DIC3, dicarboxylate carrier 3 |
0.404 |
0.102 |
| 197 |
253203_at |
AT4G34710
|
ADC2, arginine decarboxylase 2, ATADC2, SPE2 |
0.403 |
0.065 |
| 198 |
248465_at |
AT5G51200
|
EMB3142, EMBRYO DEFECTIVE 3142 |
0.402 |
0.089 |
| 199 |
246085_at |
AT5G20540
|
ATBRXL4, BREVIS RADIX-like 4, BRX-LIKE4, BRXL4, BREVIS RADIX-like 4 |
0.402 |
-0.029 |
| 200 |
256839_at |
AT3G22930
|
AtCML11, CML11, calmodulin-like 11 |
0.399 |
-0.036 |
| 201 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.399 |
-0.038 |
| 202 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
0.398 |
-0.095 |
| 203 |
245854_at |
AT5G13490
|
AAC2, ADP/ATP carrier 2 |
0.397 |
-0.021 |
| 204 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.397 |
0.014 |
| 205 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.396 |
-0.026 |
| 206 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.396 |
-0.061 |
| 207 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
0.395 |
-0.029 |
| 208 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.393 |
-0.052 |
| 209 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.393 |
-0.060 |
| 210 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.393 |
-0.121 |
| 211 |
250472_at |
AT5G10210
|
unknown |
0.393 |
-0.160 |
| 212 |
250597_at |
AT5G07680
|
ANAC079, Arabidopsis NAC domain containing protein 79, ANAC080, NAC domain containing protein 80, ATNAC4, NAC080, NAC domain containing protein 80, NAC4 |
0.392 |
-0.137 |
| 213 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.392 |
-0.108 |
| 214 |
259479_at |
AT1G19020
|
unknown |
0.392 |
-0.042 |
| 215 |
262231_at |
AT1G68740
|
PHO1;H1 |
0.391 |
-0.030 |
| 216 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.390 |
0.086 |
| 217 |
253559_at |
AT4G31140
|
[O-Glycosyl hydrolases family 17 protein] |
0.389 |
-0.011 |
| 218 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.388 |
-0.166 |
| 219 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.387 |
-0.044 |
| 220 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.387 |
-0.186 |
| 221 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
0.387 |
0.020 |
| 222 |
246682_at |
AT5G33290
|
XGD1, xylogalacturonan deficient 1 |
0.385 |
0.101 |
| 223 |
266004_at |
AT2G37330
|
ALS3, ALUMINUM SENSITIVE 3 |
0.384 |
-0.161 |
| 224 |
245977_at |
AT5G13110
|
G6PD2, glucose-6-phosphate dehydrogenase 2 |
0.384 |
0.087 |
| 225 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.383 |
-0.212 |
| 226 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.383 |
-0.107 |
| 227 |
247280_at |
AT5G64260
|
EXL2, EXORDIUM like 2 |
0.381 |
-0.039 |
| 228 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
0.381 |
-0.004 |
| 229 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.380 |
0.004 |
| 230 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.380 |
-0.078 |
| 231 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
0.379 |
-0.022 |
| 232 |
248592_at |
AT5G49280
|
[hydroxyproline-rich glycoprotein family protein] |
0.379 |
0.007 |
| 233 |
267140_at |
AT2G38250
|
[Homeodomain-like superfamily protein] |
0.378 |
-0.021 |
| 234 |
262180_at |
AT1G78050
|
PGM, phosphoglycerate/bisphosphoglycerate mutase |
0.377 |
-0.001 |
| 235 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
0.377 |
0.047 |
| 236 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
0.375 |
0.162 |
| 237 |
254043_at |
AT4G25990
|
CIL |
0.375 |
-0.048 |
| 238 |
262679_at |
AT1G75830
|
LCR67, low-molecular-weight cysteine-rich 67, PDF1.1, PLANT DEFENSIN 1.1 |
0.374 |
-0.042 |
| 239 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
0.374 |
0.049 |
| 240 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.374 |
-0.013 |
| 241 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
0.372 |
0.077 |
| 242 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
0.371 |
-0.032 |
| 243 |
263406_at |
AT2G04160
|
AIR3, AUXIN-INDUCED IN ROOT CULTURES 3 |
0.370 |
0.115 |
| 244 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.367 |
-0.124 |
| 245 |
266209_at |
AT2G27550
|
ATC, centroradialis |
0.364 |
-0.082 |
| 246 |
249427_at |
AT5G39850
|
[Ribosomal protein S4] |
0.363 |
0.272 |
| 247 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.363 |
0.182 |
| 248 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
0.362 |
-0.054 |
| 249 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.358 |
-0.036 |
| 250 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
0.356 |
-0.019 |
| 251 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
0.356 |
-0.197 |
| 252 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
0.355 |
0.056 |
| 253 |
246216_at |
AT4G36380
|
ROT3, ROTUNDIFOLIA 3 |
0.355 |
-0.148 |
| 254 |
252400_at |
AT3G48020
|
unknown |
0.355 |
-0.134 |
| 255 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.354 |
-0.018 |
| 256 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.354 |
0.113 |
| 257 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.353 |
0.053 |
| 258 |
254200_at |
AT4G24110
|
unknown |
0.353 |
-0.200 |
| 259 |
247799_at |
AT5G58840
|
[Subtilase family protein] |
0.353 |
0.035 |
| 260 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.353 |
0.190 |
| 261 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
0.352 |
-0.049 |
| 262 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.352 |
0.478 |
| 263 |
266165_at |
AT2G28190
|
CSD2, copper/zinc superoxide dismutase 2, CZSOD2, COPPER/ZINC SUPEROXIDE DISMUTASE 2 |
0.351 |
0.005 |
| 264 |
267300_at |
AT2G30140
|
UGT87A2, UDP-glucosyl transferase 87A2 |
0.351 |
0.062 |
| 265 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
0.351 |
0.013 |
| 266 |
262045_at |
AT1G80240
|
DGR1, DUF642 L-GalL responsive gene 1 |
0.351 |
-0.088 |
| 267 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.351 |
-0.181 |
| 268 |
262183_at |
AT1G77890
|
[DNA-directed RNA polymerase II protein] |
0.351 |
0.072 |
| 269 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
0.350 |
0.066 |
| 270 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.349 |
-0.220 |
| 271 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
0.349 |
0.143 |
| 272 |
261748_at |
AT1G76070
|
unknown |
0.348 |
-0.071 |
| 273 |
262098_at |
AT1G56170
|
ATHAP5B, HAP5B, NF-YC2, nuclear factor Y, subunit C2 |
0.348 |
0.095 |
| 274 |
252520_at |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
-0.678 |
-0.168 |
| 275 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
-0.630 |
-0.085 |
| 276 |
260668_at |
AT1G19530
|
unknown |
-0.623 |
-0.096 |
| 277 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.615 |
-0.107 |
| 278 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.590 |
0.056 |
| 279 |
248867_at |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
-0.526 |
0.097 |
| 280 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.513 |
-0.300 |
| 281 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.510 |
-0.227 |
| 282 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.507 |
0.081 |
| 283 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.488 |
-0.043 |
| 284 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.485 |
-0.080 |
| 285 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
-0.479 |
0.056 |
| 286 |
252149_at |
AT3G51290
|
APSR1, Altered Phosphate Starvation Response 1 |
-0.478 |
-0.089 |
| 287 |
264379_at |
AT2G25200
|
unknown |
-0.472 |
-0.143 |
| 288 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.470 |
-0.162 |
| 289 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.467 |
0.072 |
| 290 |
264501_at |
AT1G09390
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.463 |
-0.067 |
| 291 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.458 |
-0.066 |
| 292 |
250327_at |
AT5G12050
|
unknown |
-0.453 |
-0.069 |
| 293 |
250478_at |
AT5G10250
|
DOT3, DEFECTIVELY ORGANIZED TRIBUTARIES 3 |
-0.450 |
0.029 |
| 294 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.444 |
-0.201 |
| 295 |
247284_at |
AT5G64410
|
ATOPT4, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 4, OPT4, oligopeptide transporter 4 |
-0.437 |
0.057 |
| 296 |
245657_at |
AT1G56720
|
[Protein kinase superfamily protein] |
-0.436 |
0.035 |
| 297 |
250975_at |
AT5G03050
|
unknown |
-0.435 |
0.057 |
| 298 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.433 |
0.089 |
| 299 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
-0.430 |
-0.174 |
| 300 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.422 |
-0.066 |
| 301 |
260012_at |
AT1G67865
|
unknown |
-0.421 |
0.154 |
| 302 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.410 |
-0.113 |
| 303 |
262236_at |
AT1G48330
|
unknown |
-0.408 |
-0.074 |
| 304 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.406 |
-0.041 |
| 305 |
266589_at |
AT2G46250
|
[myosin heavy chain-related] |
-0.404 |
0.028 |
| 306 |
247477_at |
AT5G62340
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.398 |
-0.077 |
| 307 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
-0.397 |
-0.003 |
| 308 |
249340_at |
AT5G41140
|
[Myosin heavy chain-related protein] |
-0.395 |
0.139 |
| 309 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.390 |
-0.029 |
| 310 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.389 |
0.057 |
| 311 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
-0.388 |
-0.178 |
| 312 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.386 |
0.114 |
| 313 |
259207_at |
AT3G09050
|
unknown |
-0.384 |
0.008 |
| 314 |
253973_at |
AT4G26555
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.381 |
0.101 |
| 315 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
-0.380 |
-0.218 |
| 316 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.378 |
0.137 |
| 317 |
247977_at |
AT5G56850
|
unknown |
-0.378 |
0.135 |
| 318 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
-0.376 |
0.003 |
| 319 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.375 |
0.195 |
| 320 |
258780_at |
AT3G11910
|
AtUBP13, UBP13, ubiquitin-specific protease 13 |
-0.372 |
0.065 |
| 321 |
251750_at |
AT3G55710
|
[UDP-Glycosyltransferase superfamily protein] |
-0.372 |
-0.191 |
| 322 |
265494_at |
AT2G15680
|
AtCML30, CML30, calmodulin-like 30 |
-0.368 |
0.161 |
| 323 |
263265_at |
AT2G38820
|
unknown |
-0.361 |
-0.048 |
| 324 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
-0.357 |
-0.124 |
| 325 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.356 |
-0.035 |
| 326 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
-0.355 |
0.156 |
| 327 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
-0.353 |
0.025 |
| 328 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.353 |
0.109 |
| 329 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.350 |
0.054 |
| 330 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
-0.349 |
-0.086 |
| 331 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.347 |
-0.097 |
| 332 |
258341_at |
AT3G22790
|
NET1A, Networked 1A |
-0.340 |
0.053 |
| 333 |
265265_at |
AT2G42900
|
[Plant basic secretory protein (BSP) family protein] |
-0.339 |
-0.127 |
| 334 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.339 |
0.172 |
| 335 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.338 |
0.114 |
| 336 |
255957_at |
AT1G22160
|
unknown |
-0.338 |
0.063 |
| 337 |
261026_at |
AT1G01240
|
unknown |
-0.336 |
-0.113 |
| 338 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.336 |
0.007 |
| 339 |
247878_at |
AT5G57760
|
unknown |
-0.335 |
0.102 |
| 340 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
-0.331 |
-0.088 |
| 341 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
-0.325 |
-0.084 |
| 342 |
257381_at |
AT2G37950
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.324 |
-0.116 |
| 343 |
266324_at |
AT2G46710
|
ROPGAP3, ROP guanosine triphosphatase (GTPase)-activating protein 3 |
-0.320 |
0.120 |
| 344 |
258375_at |
AT3G17470
|
ATCRSH, CRSH, Ca2+-activated RelA/spot homolog |
-0.319 |
-0.058 |
| 345 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.317 |
0.075 |
| 346 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-0.316 |
0.110 |
| 347 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.315 |
-0.139 |
| 348 |
245353_at |
AT4G16000
|
unknown |
-0.314 |
-0.143 |
| 349 |
247118_at |
AT5G65890
|
ACR1, ACT domain repeat 1 |
-0.313 |
0.031 |
| 350 |
252143_at |
AT3G51150
|
[ATP binding microtubule motor family protein] |
-0.313 |
0.067 |
| 351 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.313 |
0.155 |
| 352 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.312 |
-0.112 |
| 353 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.312 |
0.196 |
| 354 |
267622_at |
AT2G39690
|
unknown |
-0.312 |
-0.013 |
| 355 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
-0.311 |
-0.003 |
| 356 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.311 |
-0.069 |
| 357 |
256761_at |
AT3G25670
|
[Leucine-rich repeat (LRR) family protein] |
-0.310 |
0.035 |
| 358 |
249836_at |
AT5G23420
|
HMGB6, high-mobility group box 6 |
-0.308 |
0.071 |
| 359 |
247965_at |
AT5G56540
|
AGP14, arabinogalactan protein 14, ATAGP14 |
-0.306 |
0.085 |
| 360 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.304 |
-0.128 |
| 361 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.302 |
-0.028 |
| 362 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.301 |
0.153 |
| 363 |
260968_at |
AT1G12250
|
[Pentapeptide repeat-containing protein] |
-0.299 |
-0.006 |
| 364 |
245743_at |
AT1G51080
|
unknown |
-0.299 |
0.171 |
| 365 |
263322_at |
AT2G04270
|
RNE, RNase E, RNEE/G, RNAse E/G-like |
-0.299 |
0.121 |
| 366 |
267564_at |
AT2G30740
|
[Protein kinase superfamily protein] |
-0.298 |
0.057 |
| 367 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.297 |
0.196 |
| 368 |
250744_at |
AT5G05840
|
unknown |
-0.297 |
-0.053 |
| 369 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.297 |
0.021 |
| 370 |
248688_at |
AT5G48220
|
[Aldolase-type TIM barrel family protein] |
-0.297 |
0.007 |
| 371 |
251415_at |
AT3G60380
|
unknown |
-0.297 |
0.090 |
| 372 |
263799_at |
AT2G24550
|
unknown |
-0.295 |
-0.073 |
| 373 |
267602_at |
AT2G32970
|
unknown |
-0.294 |
0.114 |
| 374 |
266831_at |
AT2G22830
|
SQE2, squalene epoxidase 2 |
-0.292 |
-0.025 |
| 375 |
249347_at |
AT5G40830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.291 |
0.263 |
| 376 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.290 |
0.162 |
| 377 |
253493_at |
AT4G31820
|
ENP, ENHANCER OF PINOID, MAB4, MACCHI-BOU 4, NPY1, NAKED PINS IN YUC MUTANTS 1 |
-0.289 |
0.135 |
| 378 |
266007_at |
AT2G37380
|
MAKR3, MEMBRANE-ASSOCIATED KINASE REGULATOR 3 |
-0.289 |
-0.007 |
| 379 |
259209_at |
AT3G09160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.288 |
-0.207 |
| 380 |
256455_at |
AT1G75190
|
unknown |
-0.287 |
0.026 |
| 381 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.287 |
0.072 |
| 382 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
-0.286 |
-0.125 |
| 383 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.286 |
-0.019 |
| 384 |
261388_at |
AT1G05385
|
LPA19, LOW PSII ACCUMULATION 19, Psb27-H1 |
-0.284 |
0.075 |
| 385 |
250751_at |
AT5G05890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.282 |
-0.087 |
| 386 |
247908_at |
AT5G57440
|
GPP2, GLYCEROL-3-PHOSPHATASE 2, GS1 |
-0.281 |
-0.084 |
| 387 |
245592_at |
AT4G14540
|
NF-YB3, nuclear factor Y, subunit B3 |
-0.281 |
0.059 |
| 388 |
248177_at |
AT5G54630
|
[zinc finger protein-related] |
-0.280 |
0.022 |
| 389 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
-0.279 |
0.070 |
| 390 |
266695_at |
AT2G19810
|
AtOZF1, AtTZF2, OZF1, Oxidation-related Zinc Finger 1, TZF2, tandem zinc finger 2 |
-0.279 |
-0.042 |
| 391 |
254221_at |
AT4G23820
|
[Pectin lyase-like superfamily protein] |
-0.279 |
0.108 |
| 392 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
-0.278 |
0.204 |
| 393 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
-0.278 |
0.143 |
| 394 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.278 |
0.013 |
| 395 |
258060_at |
AT3G26030
|
ATB' DELTA, serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B prime delta |
-0.276 |
0.081 |
| 396 |
253805_at |
AT4G28260
|
unknown |
-0.276 |
-0.111 |
| 397 |
258424_at |
AT3G16750
|
unknown |
-0.276 |
-0.063 |
| 398 |
253966_at |
AT4G26520
|
AtFBA7, FBA7, fructose-bisphosphate aldolase 7 |
-0.276 |
0.013 |
| 399 |
266825_at |
AT2G22890
|
[Kua-ubiquitin conjugating enzyme hybrid localisation domain] |
-0.276 |
-0.059 |
| 400 |
256796_at |
AT3G22210
|
unknown |
-0.275 |
-0.023 |
| 401 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.275 |
-0.124 |
| 402 |
254612_at |
AT4G19100
|
PAM68, photosynthesis affected mutant 68 |
-0.274 |
0.170 |
| 403 |
262847_at |
AT1G14840
|
ATMAP70-4, microtubule-associated proteins 70-4, MAP70-4, microtubule-associated proteins 70-4 |
-0.273 |
0.230 |
| 404 |
266946_at |
AT2G18890
|
[Protein kinase superfamily protein] |
-0.273 |
-0.045 |
| 405 |
258456_at |
AT3G22420
|
ATWNK2, ARABIDOPSIS THALIANA WITH NO K 2, WNK2, with no lysine (K) kinase 2, ZIK3 |
-0.273 |
-0.032 |
| 406 |
265297_at |
AT2G14080
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.272 |
-0.033 |
| 407 |
260004_at |
AT1G67860
|
unknown |
-0.270 |
0.216 |
| 408 |
246947_at |
AT5G25120
|
CYP71B11, ytochrome p450, family 71, subfamily B, polypeptide 11 |
-0.270 |
-0.006 |
| 409 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.270 |
0.041 |
| 410 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.270 |
0.168 |
| 411 |
246929_at |
AT5G25210
|
unknown |
-0.268 |
-0.351 |
| 412 |
257723_at |
AT3G18500
|
[DNAse I-like superfamily protein] |
-0.265 |
0.068 |
| 413 |
261608_at |
AT1G49650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.265 |
0.132 |
| 414 |
267057_at |
AT2G32500
|
[Stress responsive alpha-beta barrel domain protein] |
-0.265 |
0.020 |
| 415 |
264096_at |
AT1G78995
|
unknown |
-0.264 |
0.079 |
| 416 |
252716_at |
AT3G43920
|
ATDCL3, DICER-LIKE 3, DCL3, dicer-like 3 |
-0.264 |
0.044 |
| 417 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.264 |
-0.079 |
| 418 |
256935_at |
AT3G22570
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.263 |
-0.014 |
| 419 |
253048_at |
AT4G37560
|
[Acetamidase/Formamidase family protein] |
-0.263 |
-0.086 |
| 420 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
-0.261 |
-0.216 |
| 421 |
254362_at |
AT4G22160
|
unknown |
-0.260 |
-0.008 |
| 422 |
254369_at |
AT4G21720
|
unknown |
-0.260 |
0.117 |
| 423 |
267481_at |
AT2G02780
|
Leucine-rich repeat protein kinase family protein |
-0.259 |
0.069 |
| 424 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.259 |
0.104 |
| 425 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.259 |
-0.112 |
| 426 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.256 |
0.177 |
| 427 |
249694_at |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
-0.255 |
0.017 |
| 428 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
-0.254 |
0.091 |
| 429 |
247694_at |
AT5G59750
|
AtRIBA3, RIBA3, homolog of ribA 3 |
-0.254 |
-0.015 |
| 430 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
-0.254 |
-0.188 |
| 431 |
265085_at |
AT1G03780
|
AtTPX2, TPX2, targeting protein for XKLP2 |
-0.253 |
-0.174 |
| 432 |
265726_at |
AT2G32010
|
CVL1, CVP2 like 1 |
-0.252 |
0.001 |
| 433 |
263805_at |
AT2G40400
|
unknown |
-0.252 |
0.038 |
| 434 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.252 |
0.254 |
| 435 |
257759_at |
AT3G23070
|
ATCFM3A, CFM3A, CRM family member 3A |
-0.251 |
0.117 |
| 436 |
248853_at |
AT5G46570
|
BSK2, brassinosteroid-signaling kinase 2 |
-0.251 |
0.171 |
| 437 |
266682_at |
AT2G19780
|
[Leucine-rich repeat (LRR) family protein] |
-0.251 |
0.068 |
| 438 |
248818_at |
AT5G47040
|
LON2, lon protease 2 |
-0.250 |
0.020 |
| 439 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
-0.250 |
-0.043 |
| 440 |
261888_at |
AT1G80800
|
[pseudogene] |
-0.249 |
-0.019 |
| 441 |
253686_at |
AT4G29750
|
[CRS1 / YhbY (CRM) domain-containing protein] |
-0.247 |
0.134 |
| 442 |
250550_at |
AT5G07870
|
[HXXXD-type acyl-transferase family protein] |
-0.247 |
0.172 |
| 443 |
259448_at |
AT1G13790
|
FDM4, factor of DNA methylation 4 |
-0.246 |
0.057 |
| 444 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.245 |
0.016 |
| 445 |
255939_at |
AT1G12730
|
[GPI transamidase subunit PIG-U] |
-0.245 |
-0.076 |
| 446 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.245 |
0.140 |
| 447 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
-0.244 |
0.063 |
| 448 |
254545_at |
AT4G19830
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.244 |
-0.050 |
| 449 |
249493_at |
AT5G39080
|
[HXXXD-type acyl-transferase family protein] |
-0.242 |
0.115 |
| 450 |
253661_at |
AT4G30130
|
unknown |
-0.242 |
-0.172 |
| 451 |
257221_at |
AT3G27920
|
ATGL1, ATMYB0, myb domain protein 0, GL1, GLABRA 1, MYB0, myb domain protein 0 |
-0.242 |
0.057 |
| 452 |
257201_at |
AT3G23740
|
unknown |
-0.241 |
0.009 |
| 453 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
-0.240 |
-0.183 |
| 454 |
261512_at |
AT1G71790
|
AtCPB, CPB, capping protein B |
-0.239 |
0.062 |
| 455 |
263715_at |
AT2G20570
|
GBF's pro-rich region-interacting factor 1 |
-0.239 |
0.076 |
| 456 |
254727_at |
AT4G13670
|
PTAC5, plastid transcriptionally active 5 |
-0.239 |
-0.009 |
| 457 |
251759_at |
AT3G55630
|
ATDFD, DHFS-FPGS homolog D, DFD, DHFS-FPGS homolog D, FPGS3, folylpolyglutamate synthetase 3 |
-0.238 |
-0.083 |
| 458 |
261611_at |
AT1G49730
|
[Protein kinase superfamily protein] |
-0.238 |
0.086 |
| 459 |
254836_at |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
-0.237 |
-0.187 |
| 460 |
253495_at |
AT4G31850
|
PGR3, proton gradient regulation 3 |
-0.237 |
0.011 |
| 461 |
263163_at |
AT1G03160
|
FZL, FZO-like |
-0.236 |
0.025 |
| 462 |
253330_at |
AT4G33530
|
KUP5, K+ uptake permease 5 |
-0.236 |
0.070 |
| 463 |
252077_at |
AT3G51720
|
unknown |
-0.236 |
-0.002 |
| 464 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.235 |
-0.193 |
| 465 |
251142_at |
AT5G01015
|
unknown |
-0.235 |
-0.043 |
| 466 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
-0.235 |
0.065 |
| 467 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.234 |
-0.159 |
| 468 |
256452_at |
AT1G75240
|
AtHB33, homeobox protein 33, HB33, homeobox protein 33, ZHD5, zinc-finger homeodomain 5 |
-0.234 |
0.044 |
| 469 |
246487_at |
AT5G16030
|
unknown |
-0.233 |
-0.060 |
| 470 |
254118_at |
AT4G24790
|
[AAA-type ATPase family protein] |
-0.233 |
-0.001 |
| 471 |
246493_at |
AT5G16180
|
ATCRS1, ARABIDOPSIS ORTHOLOG OF MAIZE CHLOROPLAST SPLICING FACTOR CRS1, CRS1, ortholog of maize chloroplast splicing factor CRS1 |
-0.233 |
0.021 |
| 472 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
-0.232 |
0.038 |
| 473 |
255854_at |
AT1G67050
|
unknown |
-0.232 |
0.027 |
| 474 |
256599_at |
AT3G14760
|
unknown |
-0.232 |
-0.053 |
| 475 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
-0.231 |
0.121 |
| 476 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.231 |
-0.137 |
| 477 |
256282_at |
AT3G12550
|
FDM3, factor of DNA methylation 3 |
-0.228 |
-0.068 |
| 478 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
-0.228 |
0.030 |
| 479 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.227 |
-0.041 |
| 480 |
261801_at |
AT1G30520
|
AAE14, acyl-activating enzyme 14 |
-0.227 |
-0.033 |
| 481 |
261507_at |
AT1G71720
|
PDE338, PIGMENT DEFECTIVE 338, RLSB, RbcL mRNA S1 binding domain protein |
-0.227 |
0.018 |
| 482 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
-0.226 |
-0.243 |
| 483 |
245074_at |
AT2G23200
|
[Protein kinase superfamily protein] |
-0.226 |
0.117 |
| 484 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
-0.226 |
-0.140 |
| 485 |
247553_at |
AT5G60910
|
AGL8, AGAMOUS-like 8, FUL, FRUITFULL |
-0.226 |
-0.159 |
| 486 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.226 |
0.134 |
| 487 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.225 |
0.054 |
| 488 |
257476_at |
AT1G80960
|
[F-box and Leucine Rich Repeat domains containing protein] |
-0.225 |
-0.029 |
| 489 |
245305_at |
AT4G17215
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.225 |
-0.215 |
| 490 |
250586_at |
AT5G07630
|
[lipid transporters] |
-0.225 |
0.047 |
| 491 |
266860_at |
AT2G26870
|
NPC2, non-specific phospholipase C2 |
-0.225 |
-0.073 |
| 492 |
266089_at |
AT2G38010
|
[Neutral/alkaline non-lysosomal ceramidase] |
-0.224 |
0.129 |
| 493 |
253406_at |
AT4G32890
|
GATA9, GATA transcription factor 9 |
-0.224 |
-0.132 |
| 494 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
-0.224 |
-0.208 |
| 495 |
257815_at |
AT3G25130
|
unknown |
-0.224 |
0.039 |
| 496 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.224 |
-0.131 |
| 497 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.223 |
0.252 |
| 498 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.222 |
0.183 |
| 499 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.222 |
-0.146 |
| 500 |
264653_at |
AT1G08980
|
AMI1, amidase 1, ATAMI1, AMIDASE-LIKE PROTEIN 1, ATTOC64-I, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-I, TOC64-I, TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-I |
-0.222 |
-0.032 |
| 501 |
248402_at |
AT5G52100
|
CRR1, chlororespiration reduction 1 |
-0.222 |
0.066 |
| 502 |
262752_at |
AT1G16330
|
CYCB3;1, cyclin b3;1 |
-0.222 |
-0.200 |
| 503 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
-0.222 |
-0.176 |
| 504 |
248385_at |
AT5G51910
|
[TCP family transcription factor ] |
-0.222 |
-0.131 |
| 505 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.222 |
-0.094 |
| 506 |
259021_at |
AT3G07540
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.222 |
0.197 |
| 507 |
252433_at |
AT3G47560
|
[alpha/beta-Hydrolases superfamily protein] |
-0.222 |
0.048 |
| 508 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
-0.222 |
0.037 |
| 509 |
249709_at |
AT5G35670
|
iqd33, IQ-domain 33 |
-0.221 |
-0.052 |
| 510 |
254474_at |
AT4G20390
|
[Uncharacterised protein family (UPF0497)] |
-0.221 |
0.013 |
| 511 |
248294_at |
AT5G53060
|
RCF3, regulator of CBF gene expression 3, SHI1, SHINY 1 |
-0.220 |
0.072 |
| 512 |
264549_at |
AT1G09440
|
[Protein kinase superfamily protein] |
-0.220 |
-0.013 |
| 513 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.220 |
-0.184 |
| 514 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.219 |
0.046 |
| 515 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
-0.218 |
-0.050 |
| 516 |
259637_at |
AT1G52260
|
ATPDI3, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 3, ATPDIL1-5, PDI-like 1-5, PDI3, PROTEIN DISULFIDE ISOMERASE 3, PDIL1-5, PDI-like 1-5 |
-0.218 |
0.042 |
| 517 |
254412_at |
AT4G21430
|
B160 |
-0.218 |
-0.057 |
| 518 |
261477_at |
AT1G14310
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.217 |
-0.041 |
| 519 |
263067_at |
AT2G17550
|
TRM26, TON1 Recruiting Motif 26 |
-0.216 |
-0.054 |
| 520 |
249588_at |
AT5G37790
|
[Protein kinase superfamily protein] |
-0.216 |
-0.077 |
| 521 |
262668_at |
AT1G62830
|
ATLSD1, ARABIDOPSIS LYSINE-SPECIFIC HISTONE DEMETHYLASE, ATSWP1, LDL1, LSD1-like 1, LSD1, LYSINE-SPECIFIC HISTONE DEMETHYLASE, SWP1 |
-0.216 |
0.058 |
| 522 |
261685_at |
AT1G47290
|
3BETAHSD/D1, 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 1, AT3BETAHSD/D1, 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 1 |
-0.216 |
-0.071 |
| 523 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.216 |
-0.144 |
| 524 |
247885_at |
AT5G57830
|
unknown |
-0.215 |
-0.144 |
| 525 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
-0.215 |
-0.176 |
| 526 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
-0.215 |
0.114 |
| 527 |
258836_at |
AT3G07210
|
unknown |
-0.214 |
0.032 |
| 528 |
251605_at |
AT3G57830
|
[Leucine-rich repeat protein kinase family protein] |
-0.214 |
0.021 |
| 529 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
-0.214 |
-0.009 |
| 530 |
262086_at |
AT1G56050
|
[GTP-binding protein-related] |
-0.214 |
0.037 |
| 531 |
259394_at |
AT1G06420
|
unknown |
-0.214 |
-0.157 |
| 532 |
248584_at |
AT5G49960
|
unknown |
-0.213 |
0.093 |
| 533 |
260819_at |
AT1G06850
|
AtbZIP52, basic leucine-zipper 52, bZIP52, basic leucine-zipper 52 |
-0.213 |
0.078 |
| 534 |
257739_at |
AT3G27470
|
unknown |
-0.213 |
0.046 |
| 535 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
-0.212 |
0.178 |
| 536 |
255817_at |
AT2G33330
|
PDLP3, plasmodesmata-located protein 3 |
-0.212 |
0.115 |
| 537 |
260684_at |
AT1G17590
|
NF-YA8, nuclear factor Y, subunit A8 |
-0.212 |
-0.173 |
| 538 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
-0.212 |
0.004 |
| 539 |
264799_at |
AT1G08550
|
AVDE1, ARABIDOPSIS VIOLAXANTHIN DE-EPOXIDASE 1, NPQ1, non-photochemical quenching 1 |
-0.212 |
0.049 |
| 540 |
249993_at |
AT5G18570
|
ATOBGC, ATOBGL, OBG-like protein, CPSAR1, chloroplastic SAR1, EMB269, EMBRYO DEFECTIVE 269, EMB3138, EMBRYO DEFECTIVE 3138 |
-0.212 |
-0.059 |
| 541 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.212 |
0.198 |
| 542 |
261451_at |
AT1G21060
|
unknown |
-0.212 |
-0.088 |
| 543 |
246555_at |
AT5G15470
|
GAUT14, galacturonosyltransferase 14 |
-0.212 |
0.170 |
| 544 |
256088_at |
AT1G20810
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.211 |
-0.013 |
| 545 |
245846_at |
AT1G26130
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.211 |
0.040 |
| 546 |
245778_at |
AT1G73530
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.211 |
-0.005 |