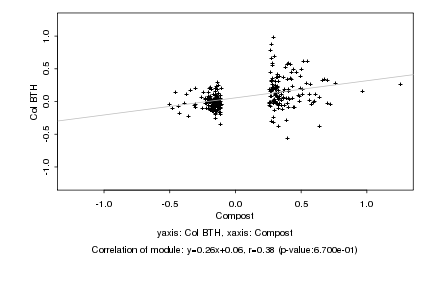
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Compost |
Log2 signal ratio
Col BTH |
| 1 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
1.253 |
0.267 |
| 2 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.966 |
0.162 |
| 3 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.758 |
0.277 |
| 4 |
265658_at |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
0.720 |
-0.038 |
| 5 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.698 |
0.330 |
| 6 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.693 |
-0.026 |
| 7 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.672 |
0.345 |
| 8 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.658 |
0.327 |
| 9 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
0.639 |
-0.379 |
| 10 |
260117_at |
AT1G33950
|
[Avirulence induced gene (AIG1) family protein] |
0.634 |
0.066 |
| 11 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.606 |
0.117 |
| 12 |
250956_at |
AT5G03210
|
AtDIP2, DIP2, DBP-interacting protein 2 |
0.598 |
0.014 |
| 13 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.587 |
-0.003 |
| 14 |
261249_at |
AT1G05880
|
ARI12, ARIADNE 12, ATARI12, ARABIDOPSIS ARIADNE 12 |
0.578 |
-0.031 |
| 15 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.567 |
0.270 |
| 16 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.561 |
0.109 |
| 17 |
249438_at |
AT5G40010
|
AATP1, AAA-ATPase 1, ASD, ATPase-in-Seed-Development |
0.556 |
0.028 |
| 18 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.548 |
0.617 |
| 19 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.537 |
0.283 |
| 20 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.517 |
0.615 |
| 21 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.510 |
0.189 |
| 22 |
245738_at |
AT1G44130
|
[Eukaryotic aspartyl protease family protein] |
0.509 |
0.115 |
| 23 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.499 |
0.494 |
| 24 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.494 |
0.201 |
| 25 |
259385_at |
AT1G13470
|
unknown |
0.493 |
0.388 |
| 26 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.487 |
0.088 |
| 27 |
257673_at |
AT3G20370
|
[TRAF-like family protein] |
0.487 |
0.010 |
| 28 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.478 |
0.094 |
| 29 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.461 |
0.445 |
| 30 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.446 |
-0.091 |
| 31 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.440 |
-0.081 |
| 32 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.440 |
0.493 |
| 33 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.433 |
0.048 |
| 34 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.430 |
0.232 |
| 35 |
246373_at |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
0.424 |
0.444 |
| 36 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
0.422 |
0.339 |
| 37 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
0.413 |
0.362 |
| 38 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.412 |
0.576 |
| 39 |
258301_at |
AT3G30510
|
transposable element gene |
0.404 |
0.037 |
| 40 |
266151_x_at |
AT2G12300
|
[CACTA-like transposase family (Ptta/En/Spm), has a 3.8e-41 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.403 |
0.052 |
| 41 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
0.403 |
0.584 |
| 42 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.402 |
-0.088 |
| 43 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.398 |
0.188 |
| 44 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.397 |
0.164 |
| 45 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.396 |
-0.559 |
| 46 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.394 |
0.345 |
| 47 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
0.393 |
0.570 |
| 48 |
265754_x_at |
AT2G10640
|
[CACTA-like transposase family (Ptta/En/Spm), has a 7.4e-42 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.390 |
-0.026 |
| 49 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.390 |
0.577 |
| 50 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.383 |
-0.278 |
| 51 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.381 |
0.048 |
| 52 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.380 |
0.524 |
| 53 |
246605_at |
AT5G35340
|
[pseudogene] |
0.367 |
-0.047 |
| 54 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.367 |
0.190 |
| 55 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.362 |
0.068 |
| 56 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.362 |
0.164 |
| 57 |
249896_at |
AT5G22530
|
unknown |
0.361 |
0.381 |
| 58 |
253378_at |
AT4G33310
|
unknown |
0.360 |
0.049 |
| 59 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.355 |
-0.110 |
| 60 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.349 |
0.113 |
| 61 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.348 |
-0.046 |
| 62 |
252007_at |
AT3G52830
|
unknown |
0.346 |
0.019 |
| 63 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.335 |
0.092 |
| 64 |
257418_at |
AT1G30850
|
RSH4, root hair specific 4 |
0.331 |
-0.018 |
| 65 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.329 |
0.387 |
| 66 |
264832_at |
AT1G03660
|
[Ankyrin-repeat containing protein] |
0.329 |
0.051 |
| 67 |
262452_at |
AT1G11210
|
unknown |
0.327 |
-0.048 |
| 68 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
0.325 |
0.139 |
| 69 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.325 |
0.315 |
| 70 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.325 |
0.181 |
| 71 |
246855_at |
AT5G26280
|
[TRAF-like family protein] |
0.324 |
0.032 |
| 72 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
0.324 |
-0.099 |
| 73 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.323 |
-0.376 |
| 74 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.320 |
0.426 |
| 75 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.320 |
0.216 |
| 76 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
0.318 |
-0.034 |
| 77 |
261366_at |
AT1G53100
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.318 |
0.232 |
| 78 |
253504_at |
AT4G31960
|
unknown |
0.317 |
-0.022 |
| 79 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.316 |
0.370 |
| 80 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.316 |
0.070 |
| 81 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.311 |
0.147 |
| 82 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
0.308 |
0.095 |
| 83 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
0.305 |
0.380 |
| 84 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
0.304 |
-0.012 |
| 85 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.303 |
0.137 |
| 86 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.303 |
0.107 |
| 87 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.302 |
-0.011 |
| 88 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.301 |
0.115 |
| 89 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.301 |
0.245 |
| 90 |
256569_at |
AT3G19550
|
unknown |
0.301 |
0.096 |
| 91 |
257154_at |
AT3G27210
|
unknown |
0.300 |
0.192 |
| 92 |
254229_at |
AT4G23610
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.300 |
0.307 |
| 93 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
0.300 |
0.015 |
| 94 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.298 |
0.212 |
| 95 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.296 |
0.699 |
| 96 |
262942_at |
AT1G79450
|
ALIS5, ALA-interacting subunit 5 |
0.296 |
0.200 |
| 97 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
0.295 |
-0.126 |
| 98 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
0.295 |
0.038 |
| 99 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.289 |
-0.230 |
| 100 |
256840_x_at |
AT3G32000
|
[gypsy-like retrotransposon family (Athila), has a 5.4e-305 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
0.289 |
0.022 |
| 101 |
263293_x_at |
AT2G10880
|
[hypothetical protein, similar to hypothetical protein GB:AAC26673] |
0.289 |
0.012 |
| 102 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
0.286 |
-0.310 |
| 103 |
261960_at |
AT1G36550
|
[similar to retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa (japonica cultivar-group)] (GB:ABA98367.2)] |
0.285 |
-0.015 |
| 104 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.285 |
0.218 |
| 105 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.284 |
0.197 |
| 106 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.284 |
0.980 |
| 107 |
246001_at |
AT5G20790
|
unknown |
0.282 |
0.013 |
| 108 |
251419_at |
AT3G60470
|
unknown |
0.279 |
0.553 |
| 109 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.279 |
0.357 |
| 110 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
0.278 |
0.324 |
| 111 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.277 |
0.195 |
| 112 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
0.275 |
0.227 |
| 113 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.275 |
0.274 |
| 114 |
255406_at |
AT4G03450
|
[Ankyrin repeat family protein] |
0.275 |
0.209 |
| 115 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.275 |
0.583 |
| 116 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.274 |
0.876 |
| 117 |
252345_at |
AT3G48640
|
unknown |
0.274 |
0.156 |
| 118 |
254091_at |
AT4G25070
|
unknown |
0.272 |
0.334 |
| 119 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.272 |
0.318 |
| 120 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.271 |
0.658 |
| 121 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.268 |
0.089 |
| 122 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
0.267 |
-0.302 |
| 123 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.266 |
-0.061 |
| 124 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.265 |
0.453 |
| 125 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.265 |
-0.008 |
| 126 |
255912_at |
AT1G66960
|
LUP5 |
0.263 |
0.788 |
| 127 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.263 |
-0.008 |
| 128 |
266376_at |
AT2G14620
|
XTH10, xyloglucan endotransglucosylase/hydrolase 10 |
0.262 |
0.083 |
| 129 |
260943_at |
AT1G45145
|
ATH5, THIOREDOXIN H-TYPE 5, ATTRX5, thioredoxin H-type 5, LIV1, LOCUS OF INSENSITIVITY TO VICTORIN 1, TRX-h5, THIOREDOXIN H-TYPE 5, TRX5, thioredoxin H-type 5 |
0.260 |
0.160 |
| 130 |
257335_at |
ATMG01400
|
ORF105B |
0.259 |
-0.019 |
| 131 |
260713_at |
AT1G17615
|
[Disease resistance protein (TIR-NBS class)] |
0.258 |
-0.011 |
| 132 |
249454_at |
AT5G39520
|
unknown |
0.258 |
0.186 |
| 133 |
256748_x_at |
AT3G30396
|
[CACTA-like transposase family (Ptta/En/Spm), has a 3.1e-39 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.258 |
-0.047 |
| 134 |
267459_at |
AT2G33850
|
unknown |
-0.507 |
-0.045 |
| 135 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.485 |
-0.101 |
| 136 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.462 |
0.147 |
| 137 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.434 |
-0.062 |
| 138 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.427 |
-0.173 |
| 139 |
253056_at |
AT4G37650
|
SGR7, SHOOT GRAVITROPISM 7, SHR, SHORT ROOT |
-0.392 |
-0.026 |
| 140 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-0.377 |
0.115 |
| 141 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.363 |
-0.215 |
| 142 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.344 |
0.176 |
| 143 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
-0.317 |
-0.057 |
| 144 |
247005_at |
AT5G67520
|
adenosine-5'-phosphosulfate (APS) kinase 4 |
-0.311 |
0.199 |
| 145 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
-0.311 |
-0.038 |
| 146 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.305 |
-0.084 |
| 147 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
-0.265 |
0.076 |
| 148 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.252 |
-0.105 |
| 149 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.249 |
0.152 |
| 150 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.234 |
-0.021 |
| 151 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-0.234 |
0.049 |
| 152 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.228 |
0.043 |
| 153 |
250025_at |
AT5G18290
|
SIP1;2, SIP1B, SMALL AND BASIC INTRINSIC PROTEIN 1B |
-0.223 |
-0.101 |
| 154 |
252820_at |
AT3G42640
|
AHA8, H(+)-ATPase 8, HA8, H(+)-ATPase 8 |
-0.221 |
-0.022 |
| 155 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
-0.220 |
0.054 |
| 156 |
259346_at |
AT3G03910
|
glutamate dehydrogenase 3 |
-0.213 |
0.008 |
| 157 |
246781_at |
AT5G27350
|
SFP1 |
-0.212 |
0.032 |
| 158 |
259685_at |
AT1G63090
|
AtPP2-A11, phloem protein 2-A11, PP2-A11, phloem protein 2-A11 |
-0.212 |
0.141 |
| 159 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
-0.210 |
-0.096 |
| 160 |
261106_at |
AT1G62990
|
IXR11, KNAT7, KNOTTED-like homeobox of Arabidopsis thaliana 7 |
-0.208 |
-0.058 |
| 161 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
-0.208 |
0.089 |
| 162 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
-0.205 |
0.015 |
| 163 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
-0.204 |
0.212 |
| 164 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.203 |
-0.114 |
| 165 |
247361_at |
AT5G63480
|
unknown |
-0.201 |
0.068 |
| 166 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.199 |
-0.007 |
| 167 |
245934_at |
AT5G09330
|
ANAC082, NAC domain containing protein 82, NAC082, NAC domain containing protein 82, VNI1, VND-interacting 1 |
-0.197 |
0.091 |
| 168 |
247719_at |
AT5G59305
|
unknown |
-0.196 |
0.038 |
| 169 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.196 |
0.227 |
| 170 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
-0.195 |
-0.136 |
| 171 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.193 |
-0.004 |
| 172 |
247903_at |
AT5G57340
|
unknown |
-0.191 |
-0.120 |
| 173 |
261124_at |
AT1G04900
|
unknown |
-0.189 |
0.036 |
| 174 |
251109_at |
AT5G01600
|
ATFER1, ARABIDOPSIS THALIANA FERRETIN 1, FER1, ferretin 1 |
-0.188 |
-0.113 |
| 175 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.188 |
-0.065 |
| 176 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
-0.187 |
0.002 |
| 177 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
-0.186 |
-0.099 |
| 178 |
266015_at |
AT2G24190
|
SDR2, short-chain dehydrogenase/reductase 2 |
-0.185 |
-0.067 |
| 179 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.184 |
-0.052 |
| 180 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
-0.184 |
0.196 |
| 181 |
248385_at |
AT5G51910
|
[TCP family transcription factor ] |
-0.181 |
-0.048 |
| 182 |
265412_at |
AT2G16640
|
ATTOC132, MULTIMERIC TRANSLOCON COMPLEX IN THE OUTER ENVELOPE MEMBRANE 132, TOC132, multimeric translocon complex in the outer envelope membrane 132 |
-0.181 |
0.022 |
| 183 |
264341_at |
AT1G70270
|
unknown |
-0.180 |
-0.144 |
| 184 |
266106_at |
AT2G45170
|
ATATG8E, AUTOPHAGY 8E, ATG8E, AUTOPHAGY 8E |
-0.173 |
0.118 |
| 185 |
257132_at |
AT3G20230
|
[Ribosomal L18p/L5e family protein] |
-0.173 |
-0.052 |
| 186 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
-0.173 |
-0.144 |
| 187 |
267260_at |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
-0.172 |
-0.083 |
| 188 |
258030_at |
AT3G27590
|
unknown |
-0.172 |
-0.004 |
| 189 |
251722_at |
AT3G56200
|
[Transmembrane amino acid transporter family protein] |
-0.172 |
-0.028 |
| 190 |
263788_at |
AT2G24580
|
[FAD-dependent oxidoreductase family protein] |
-0.171 |
-0.054 |
| 191 |
256096_at |
AT1G13650
|
unknown |
-0.167 |
0.044 |
| 192 |
245884_at |
AT5G09300
|
[Thiamin diphosphate-binding fold (THDP-binding) superfamily protein] |
-0.166 |
-0.030 |
| 193 |
245912_at |
AT5G19600
|
SULTR3;5, sulfate transporter 3;5 |
-0.165 |
-0.013 |
| 194 |
262071_at |
AT1G59510
|
CF9 |
-0.163 |
-0.114 |
| 195 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.163 |
-0.081 |
| 196 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
-0.162 |
0.039 |
| 197 |
261727_at |
AT1G76090
|
SMT3, sterol methyltransferase 3 |
-0.161 |
0.038 |
| 198 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.159 |
0.121 |
| 199 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
-0.159 |
-0.169 |
| 200 |
264826_at |
AT1G03410
|
2A6 |
-0.158 |
-0.021 |
| 201 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.158 |
-0.245 |
| 202 |
253195_at |
AT4G35420
|
DRL1, dihydroflavonol 4-reductase-like1, TKPR1, tetraketide alpha-pyrone reductase 1 |
-0.156 |
0.042 |
| 203 |
247609_at |
AT5G60940
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.155 |
0.000 |
| 204 |
249427_at |
AT5G39850
|
[Ribosomal protein S4] |
-0.155 |
0.098 |
| 205 |
264590_at |
AT2G17710
|
unknown |
-0.155 |
0.212 |
| 206 |
264692_at |
AT1G70000
|
[myb-like transcription factor family protein] |
-0.155 |
0.162 |
| 207 |
257547_at |
AT3G13000
|
unknown |
-0.154 |
-0.198 |
| 208 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.153 |
-0.107 |
| 209 |
265005_at |
AT1G61667
|
unknown |
-0.153 |
0.103 |
| 210 |
247754_at |
AT5G59080
|
unknown |
-0.152 |
0.014 |
| 211 |
256025_at |
AT1G58370
|
ATXYN1, ARABIDOPSIS THALIANA XYLANASE 1, RXF12 |
-0.152 |
0.022 |
| 212 |
261667_at |
AT1G18460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.152 |
-0.026 |
| 213 |
250189_at |
AT5G14410
|
unknown |
-0.151 |
-0.138 |
| 214 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.150 |
0.106 |
| 215 |
247049_at |
AT5G66440
|
unknown |
-0.148 |
-0.047 |
| 216 |
255957_at |
AT1G22160
|
unknown |
-0.148 |
-0.076 |
| 217 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-0.147 |
0.232 |
| 218 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
-0.147 |
-0.012 |
| 219 |
265709_at |
AT2G03540
|
unknown |
-0.147 |
0.066 |
| 220 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.146 |
0.080 |
| 221 |
256043_at |
AT1G07210
|
[Ribosomal protein S18] |
-0.145 |
-0.019 |
| 222 |
250550_at |
AT5G07870
|
[HXXXD-type acyl-transferase family protein] |
-0.144 |
0.020 |
| 223 |
261710_at |
AT1G32730
|
unknown |
-0.144 |
-0.133 |
| 224 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.144 |
0.303 |
| 225 |
248984_at |
AT5G45140
|
NRPC2, nuclear RNA polymerase C2 |
-0.143 |
-0.005 |
| 226 |
266516_at |
AT2G47880
|
[Glutaredoxin family protein] |
-0.142 |
0.074 |
| 227 |
254573_at |
AT4G19420
|
[Pectinacetylesterase family protein] |
-0.140 |
0.194 |
| 228 |
259207_at |
AT3G09050
|
unknown |
-0.139 |
-0.062 |
| 229 |
246860_at |
AT5G25840
|
unknown |
-0.139 |
-0.110 |
| 230 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.138 |
-0.080 |
| 231 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.138 |
0.079 |
| 232 |
248225_at |
AT5G53740
|
unknown |
-0.138 |
0.014 |
| 233 |
256920_at |
AT3G18980
|
ETP1, EIN2 targeting protein1 |
-0.137 |
0.052 |
| 234 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.136 |
0.126 |
| 235 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.135 |
0.247 |
| 236 |
259429_at |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
-0.135 |
-0.034 |
| 237 |
265243_at |
AT2G43040
|
NPG1, no pollen germination 1 |
-0.133 |
-0.059 |
| 238 |
266358_at |
AT2G32280
|
VCC, VASCULATURE COMPLEXITY AND CONNECTIVITY |
-0.133 |
0.036 |
| 239 |
259126_at |
AT3G02280
|
AtTAH18, TAH18 |
-0.131 |
0.004 |
| 240 |
247504_at |
AT5G61990
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.131 |
-0.046 |
| 241 |
248046_at |
AT5G56040
|
SKM2, STERILITY-REGULATING KINASE MEMBER 2 |
-0.130 |
-0.109 |
| 242 |
257530_at |
AT3G03040
|
[F-box/RNI-like superfamily protein] |
-0.129 |
0.059 |
| 243 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
-0.128 |
-0.016 |
| 244 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
-0.126 |
-0.036 |
| 245 |
254503_at |
AT4G20010
|
OSB2, ORGANELLAR SINGLE-STRANDED DNA BINDING PROTEIN 2, PTAC9, plastid transcriptionally active 9 |
-0.126 |
0.003 |
| 246 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.124 |
-0.060 |
| 247 |
245547_at |
AT4G15300
|
CYP702A2, cytochrome P450, family 702, subfamily A, polypeptide 2 |
-0.123 |
-0.006 |
| 248 |
264830_at |
AT1G03710
|
[Cystatin/monellin superfamily protein] |
-0.123 |
-0.155 |
| 249 |
266275_at |
AT2G29370
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.122 |
-0.057 |
| 250 |
246043_at |
AT5G19380
|
CLT1, CRT (chloroquine-resistance transporter)-like transporter 1 |
-0.122 |
-0.016 |
| 251 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.122 |
-0.043 |
| 252 |
258040_at |
AT3G21190
|
AtMSR1, MSR1, Mannan Synthesis Related 1 |
-0.121 |
-0.086 |
| 253 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.119 |
-0.346 |
| 254 |
263084_at |
AT2G27180
|
unknown |
-0.119 |
0.047 |
| 255 |
262558_at |
AT1G31335
|
unknown |
-0.117 |
-0.050 |
| 256 |
266869_at |
AT2G44660
|
[ALG6, ALG8 glycosyltransferase family] |
-0.117 |
0.096 |
| 257 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.117 |
-0.157 |
| 258 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
-0.117 |
0.032 |
| 259 |
260194_at |
AT1G67530
|
[ARM repeat superfamily protein] |
-0.117 |
-0.093 |
| 260 |
248454_at |
AT5G51350
|
MOL1, MORE LATERAL GROWTH1 |
-0.116 |
-0.058 |
| 261 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.114 |
0.005 |
| 262 |
247693_at |
AT5G59730
|
ATEXO70H7, exocyst subunit exo70 family protein H7, EXO70H7, exocyst subunit exo70 family protein H7 |
-0.114 |
-0.191 |
| 263 |
252000_at |
AT3G52710
|
unknown |
-0.113 |
0.202 |
| 264 |
245462_at |
AT4G17020
|
[transcription factor-related] |
-0.113 |
-0.034 |
| 265 |
260442_at |
AT1G68220
|
unknown |
-0.113 |
-0.040 |