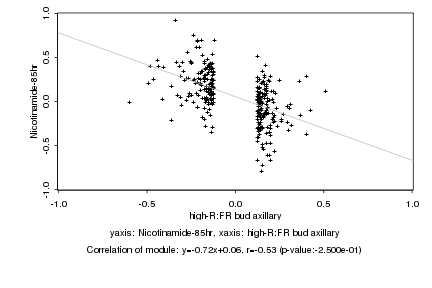
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
high-R:FR bud axillary |
Log2 signal ratio
Nicotinamide-85hr |
| 1 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
0.507 |
0.125 |
| 2 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
0.421 |
-0.100 |
| 3 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
0.400 |
-0.365 |
| 4 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
0.397 |
0.286 |
| 5 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
0.364 |
-0.157 |
| 6 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
0.362 |
0.233 |
| 7 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
0.312 |
-0.270 |
| 8 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
0.311 |
-0.032 |
| 9 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
0.305 |
-0.072 |
| 10 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
0.299 |
-0.321 |
| 11 |
252661_at |
AT3G44450
|
unknown |
0.293 |
-0.056 |
| 12 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.289 |
-0.233 |
| 13 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
0.267 |
-0.145 |
| 14 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.258 |
-0.225 |
| 15 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
0.257 |
-0.201 |
| 16 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
0.249 |
0.245 |
| 17 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
0.234 |
-0.281 |
| 18 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
0.231 |
-0.078 |
| 19 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.226 |
0.101 |
| 20 |
256527_at |
AT1G66100
|
[Plant thionin] |
0.221 |
-0.568 |
| 21 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
0.219 |
-0.154 |
| 22 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.217 |
-0.223 |
| 23 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.215 |
-0.181 |
| 24 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
0.214 |
-0.228 |
| 25 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
0.212 |
-0.006 |
| 26 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
0.211 |
0.114 |
| 27 |
246001_at |
AT5G20790
|
unknown |
0.208 |
-0.196 |
| 28 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
0.206 |
0.024 |
| 29 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
0.205 |
-0.132 |
| 30 |
250907_at |
AT5G03670
|
TRM28, TON1 Recruiting Motif 28 |
0.199 |
0.115 |
| 31 |
246494_at |
AT5G16190
|
ATCSLA11, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A11, CSLA11, cellulose synthase like A11 |
0.198 |
-0.265 |
| 32 |
257875_at |
AT3G17120
|
unknown |
0.198 |
-0.300 |
| 33 |
263236_at |
AT1G10470
|
ARR4, response regulator 4, ATRR1, RESPONCE REGULATOR 1, IBC7, INDUCED BY CYTOKININ 7, MEE7, maternal effect embryo arrest 7 |
0.197 |
0.285 |
| 34 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
0.195 |
-0.380 |
| 35 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.194 |
-0.469 |
| 36 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
0.194 |
-0.668 |
| 37 |
245465_at |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
0.192 |
0.228 |
| 38 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
0.192 |
-0.352 |
| 39 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
0.190 |
-0.341 |
| 40 |
258560_at |
AT3G06020
|
FAF4, FANTASTIC FOUR 4 |
0.190 |
0.021 |
| 41 |
252365_at |
AT3G48350
|
CEP3, cysteine endopeptidase 3 |
0.188 |
-0.612 |
| 42 |
266253_at |
AT2G27840
|
HD2D, histone deacetylase 2D, HDA13, HISTONE DEACETYLASE 13, HDT04, HDT4 |
0.187 |
-0.128 |
| 43 |
258468_at |
AT3G06070
|
unknown |
0.183 |
0.249 |
| 44 |
259680_at |
AT1G77690
|
LAX3, like AUX1 3 |
0.181 |
0.007 |
| 45 |
258965_at |
AT3G10530
|
[Transducin/WD40 repeat-like superfamily protein] |
0.181 |
-0.069 |
| 46 |
264000_at |
AT2G22500
|
ATPUMP5, PLANT UNCOUPLING MITOCHONDRIAL PROTEIN 5, DIC1, DICARBOXYLATE CARRIER 1, UCP5, uncoupling protein 5 |
0.180 |
-0.062 |
| 47 |
261772_at |
AT1G76240
|
unknown |
0.179 |
0.045 |
| 48 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.179 |
-0.608 |
| 49 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
0.179 |
-0.286 |
| 50 |
266124_at |
AT2G45080
|
cycp3;1, cyclin p3;1 |
0.175 |
0.134 |
| 51 |
265730_at |
AT2G32220
|
[Ribosomal L27e protein family] |
0.174 |
0.115 |
| 52 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.174 |
-0.093 |
| 53 |
253534_at |
AT4G31500
|
ATR4, ALTERED TRYPTOPHAN REGULATION 4, CYP83B1, cytochrome P450, family 83, subfamily B, polypeptide 1, RED1, RED ELONGATED 1, RNT1, RUNT 1, SUR2, SUPERROOT 2 |
0.174 |
0.181 |
| 54 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
0.174 |
0.197 |
| 55 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.173 |
-0.353 |
| 56 |
253252_at |
AT4G34740
|
ASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATPURF2, CIA1, CHLOROPLAST IMPORT APPARATUS 1, DOV1, DIFFERENTIAL DEVELOPMENT OF VASCULAR ASSOCIATED CELLS |
0.173 |
0.156 |
| 57 |
258470_at |
AT3G06035
|
[Glycoprotein membrane precursor GPI-anchored] |
0.172 |
-0.472 |
| 58 |
258562_at |
AT3G05980
|
unknown |
0.172 |
-0.115 |
| 59 |
247739_at |
AT5G59240
|
[Ribosomal protein S8e family protein] |
0.171 |
-0.140 |
| 60 |
250045_at |
AT5G17700
|
[MATE efflux family protein] |
0.171 |
0.101 |
| 61 |
248865_at |
AT5G46790
|
PYL1, PYR1-like 1, RCAR12, regulatory components of ABA receptor 12 |
0.171 |
-0.026 |
| 62 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
0.169 |
0.075 |
| 63 |
246200_at |
AT4G37240
|
unknown |
0.167 |
0.417 |
| 64 |
256924_at |
AT3G29590
|
AT5MAT |
0.166 |
-0.122 |
| 65 |
264895_at |
AT1G23100
|
[GroES-like family protein] |
0.163 |
0.080 |
| 66 |
260143_at |
AT1G71880
|
ATSUC1, ARABIDOPSIS THALIANA SUCROSE-PROTON SYMPORTER 1, SUC1, sucrose-proton symporter 1 |
0.163 |
0.305 |
| 67 |
260768_at |
AT1G49245
|
[Prefoldin chaperone subunit family protein] |
0.162 |
0.140 |
| 68 |
250413_at |
AT5G11160
|
APT5, adenine phosphoribosyltransferase 5 |
0.161 |
0.039 |
| 69 |
261266_at |
AT1G26770
|
AT-EXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXPA10, expansin A10, ATHEXP ALPHA 1.1, ARABIDOPSIS THALIANA EXPANSIN ALPHA 1.1, EXP10, EXPANSIN 10, EXPA10, expansin A10 |
0.160 |
0.274 |
| 70 |
265147_at |
AT1G51380
|
[DEA(D/H)-box RNA helicase family protein] |
0.160 |
0.203 |
| 71 |
255856_at |
AT1G66940
|
[protein kinase-related] |
0.159 |
-0.131 |
| 72 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.158 |
-0.159 |
| 73 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.158 |
0.234 |
| 74 |
259985_at |
AT1G76620
|
unknown |
0.157 |
-0.178 |
| 75 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
0.156 |
-0.537 |
| 76 |
257571_at |
AT3G16870
|
GATA17, GATA transcription factor 17 |
0.154 |
-0.156 |
| 77 |
249497_at |
AT5G39220
|
[alpha/beta-Hydrolases superfamily protein] |
0.151 |
0.063 |
| 78 |
251646_at |
AT3G57780
|
unknown |
0.151 |
-0.304 |
| 79 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
0.150 |
-0.514 |
| 80 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
0.149 |
-0.724 |
| 81 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.149 |
-0.020 |
| 82 |
251476_at |
AT3G59670
|
unknown |
0.149 |
-0.478 |
| 83 |
260484_at |
AT1G68360
|
[C2H2 and C2HC zinc fingers superfamily protein] |
0.149 |
0.352 |
| 84 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
0.148 |
-0.287 |
| 85 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
0.148 |
0.093 |
| 86 |
251611_at |
AT3G57940
|
unknown |
0.148 |
0.218 |
| 87 |
267177_at |
AT2G37580
|
[RING/U-box superfamily protein] |
0.147 |
0.208 |
| 88 |
250161_at |
AT5G15240
|
[Transmembrane amino acid transporter family protein] |
0.147 |
-0.007 |
| 89 |
267309_at |
AT2G19385
|
[zinc ion binding] |
0.145 |
-0.074 |
| 90 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.144 |
-0.792 |
| 91 |
253165_at |
AT4G35320
|
unknown |
0.144 |
-0.100 |
| 92 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.144 |
-0.197 |
| 93 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.144 |
0.008 |
| 94 |
246765_at |
AT5G27330
|
[Prefoldin chaperone subunit family protein] |
0.142 |
-0.288 |
| 95 |
258202_at |
AT3G13940
|
[DNA binding] |
0.141 |
0.105 |
| 96 |
261926_at |
AT1G22530
|
PATL2, PATELLIN 2 |
0.140 |
0.110 |
| 97 |
251292_at |
AT3G61920
|
unknown |
0.140 |
-0.042 |
| 98 |
257201_at |
AT3G23740
|
unknown |
0.139 |
-0.173 |
| 99 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
0.139 |
-0.129 |
| 100 |
246797_at |
AT5G26790
|
unknown |
0.139 |
-0.328 |
| 101 |
256077_at |
AT1G18090
|
[5'-3' exonuclease family protein] |
0.138 |
-0.228 |
| 102 |
251174_at |
AT3G63200
|
PLA IIIB, PLP9, PATATIN-like protein 9 |
0.138 |
-0.084 |
| 103 |
263355_at |
AT2G22100
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.138 |
0.031 |
| 104 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
0.137 |
-0.286 |
| 105 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
0.137 |
-0.141 |
| 106 |
254080_at |
AT4G25630
|
ATFIB2, FIB2, fibrillarin 2 |
0.137 |
0.057 |
| 107 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
0.136 |
-0.322 |
| 108 |
251538_at |
AT3G58660
|
[Ribosomal protein L1p/L10e family] |
0.136 |
-0.030 |
| 109 |
263047_at |
AT2G17630
|
PSAT2, phosphoserine aminotransferase 2 |
0.136 |
-0.259 |
| 110 |
260948_at |
AT1G06100
|
[Fatty acid desaturase family protein] |
0.136 |
0.092 |
| 111 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
0.136 |
-0.309 |
| 112 |
257136_at |
AT3G12890
|
ASML2, activator of spomin::LUC2 |
0.134 |
0.032 |
| 113 |
254639_at |
AT4G18750
|
DOT4, DEFECTIVELY ORGANIZED TRIBUTARIES 4 |
0.134 |
-0.134 |
| 114 |
246395_at |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.134 |
0.150 |
| 115 |
248582_at |
AT5G49910
|
cpHsc70-2, chloroplast heat shock protein 70-2, HSC70-7, HEAT SHOCK PROTEIN 70-7 |
0.133 |
0.168 |
| 116 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
0.133 |
0.021 |
| 117 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
0.133 |
-0.017 |
| 118 |
245254_at |
AT4G14680
|
APS3 |
0.132 |
0.020 |
| 119 |
266510_at |
AT2G47990
|
EDA13, EMBRYO SAC DEVELOPMENT ARREST 13, EDA19, EMBRYO SAC DEVELOPMENT ARREST 19, SWA1, SLOW WALKER1 |
0.132 |
0.254 |
| 120 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.132 |
-0.371 |
| 121 |
248372_at |
AT5G51850
|
TRM24, TON1 Recruiting Motif 24 |
0.132 |
-0.154 |
| 122 |
266770_at |
AT2G03090
|
ATEXP15, ATEXPA15, expansin A15, ATHEXP ALPHA 1.3, EXP15, EXPANSIN 15, EXPA15, expansin A15 |
0.131 |
0.068 |
| 123 |
261261_at |
AT1G26730
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.131 |
0.168 |
| 124 |
257968_at |
AT3G27550
|
[RNA-binding CRS1 / YhbY (CRM) domain protein] |
0.131 |
0.039 |
| 125 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
0.130 |
0.026 |
| 126 |
249718_at |
AT5G35740
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.130 |
-0.233 |
| 127 |
258979_at |
AT3G09440
|
[Heat shock protein 70 (Hsp 70) family protein] |
0.130 |
0.057 |
| 128 |
259020_at |
AT3G07470
|
unknown |
0.129 |
-0.007 |
| 129 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
0.129 |
0.062 |
| 130 |
248357_at |
AT5G52380
|
[VASCULAR-RELATED NAC-DOMAIN 6] |
0.129 |
-0.128 |
| 131 |
263382_at |
AT2G40230
|
[HXXXD-type acyl-transferase family protein] |
0.129 |
0.083 |
| 132 |
253578_at |
AT4G30340
|
ATDGK7, diacylglycerol kinase 7, DGK7, diacylglycerol kinase 7 |
0.128 |
-0.336 |
| 133 |
251638_at |
AT3G57490
|
[Ribosomal protein S5 family protein] |
0.128 |
0.151 |
| 134 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
0.128 |
-0.401 |
| 135 |
252133_at |
AT3G50900
|
unknown |
0.127 |
-0.056 |
| 136 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
0.127 |
-0.098 |
| 137 |
267112_at |
AT2G14750
|
AKN1, APS KINASE 1, APK, APS kinase, APK1, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 1, ATAKN1 |
0.126 |
0.066 |
| 138 |
249347_at |
AT5G40830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.126 |
-0.293 |
| 139 |
258529_at |
AT3G06740
|
GATA15, GATA transcription factor 15 |
0.126 |
-0.290 |
| 140 |
248061_at |
AT5G55340
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
0.125 |
-0.032 |
| 141 |
260395_at |
AT1G69780
|
ATHB13 |
0.125 |
-0.100 |
| 142 |
248954_at |
AT5G45420
|
maMYB, membrane anchored MYB |
0.125 |
0.086 |
| 143 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
0.124 |
0.517 |
| 144 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
0.124 |
0.164 |
| 145 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.124 |
-0.451 |
| 146 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
0.124 |
0.031 |
| 147 |
261728_at |
AT1G76160
|
sks5, SKU5 similar 5 |
0.124 |
0.123 |
| 148 |
254789_at |
AT4G12880
|
AtENODL19, ENODL19, early nodulin-like protein 19 |
0.124 |
-0.113 |
| 149 |
260824_at |
AT1G06720
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.123 |
-0.400 |
| 150 |
258316_at |
AT3G22660
|
[rRNA processing protein-related] |
0.123 |
0.061 |
| 151 |
255887_at |
AT1G20370
|
[Pseudouridine synthase family protein] |
0.123 |
-0.166 |
| 152 |
267474_at |
AT2G02740
|
ATWHY3, A. THALIANA WHIRLY 3, PTAC11, PLASTID TRANSCRIPTIONALLY ACTIVE11, WHY3, WHIRLY 3 |
0.123 |
0.017 |
| 153 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
0.123 |
-0.663 |
| 154 |
262745_at |
AT1G28600
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.122 |
0.282 |
| 155 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
0.121 |
0.029 |
| 156 |
260494_at |
AT2G41820
|
PXC3, PXY/TDR-correlated 3 |
0.121 |
-0.302 |
| 157 |
258089_at |
AT3G14740
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.121 |
-0.203 |
| 158 |
252952_at |
AT4G38710
|
[glycine-rich protein] |
0.120 |
0.234 |
| 159 |
251012_at |
AT5G02580
|
unknown |
-0.603 |
-0.000 |
| 160 |
248509_at |
AT5G50335
|
unknown |
-0.494 |
0.213 |
| 161 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.482 |
0.409 |
| 162 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
-0.469 |
0.255 |
| 163 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-0.443 |
0.467 |
| 164 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.436 |
0.408 |
| 165 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.417 |
0.025 |
| 166 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
-0.410 |
0.391 |
| 167 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
-0.364 |
0.174 |
| 168 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.364 |
-0.211 |
| 169 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
-0.341 |
0.932 |
| 170 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.339 |
0.446 |
| 171 |
253152_at |
AT4G35690
|
unknown |
-0.329 |
0.073 |
| 172 |
264580_at |
AT1G05340
|
unknown |
-0.321 |
0.409 |
| 173 |
264494_at |
AT1G27461
|
unknown |
-0.316 |
0.288 |
| 174 |
247878_at |
AT5G57760
|
unknown |
-0.312 |
0.050 |
| 175 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.307 |
-0.038 |
| 176 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.303 |
0.447 |
| 177 |
265539_at |
AT2G15830
|
unknown |
-0.296 |
0.351 |
| 178 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
-0.290 |
0.243 |
| 179 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.278 |
0.021 |
| 180 |
258327_at |
AT3G22640
|
PAP85 |
-0.278 |
0.265 |
| 181 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.276 |
0.558 |
| 182 |
265387_at |
AT2G20670
|
unknown |
-0.269 |
0.271 |
| 183 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
-0.269 |
0.068 |
| 184 |
254954_at |
AT4G10910
|
unknown |
-0.263 |
0.100 |
| 185 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
-0.258 |
0.463 |
| 186 |
263169_at |
AT1G03010
|
[Phototropic-responsive NPH3 family protein] |
-0.254 |
0.076 |
| 187 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
-0.254 |
0.437 |
| 188 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
-0.253 |
0.451 |
| 189 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
-0.242 |
0.757 |
| 190 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
-0.242 |
0.245 |
| 191 |
249941_at |
AT5G22270
|
unknown |
-0.240 |
-0.003 |
| 192 |
250937_at |
AT5G03230
|
unknown |
-0.234 |
0.252 |
| 193 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.228 |
0.206 |
| 194 |
265276_at |
AT2G28400
|
unknown |
-0.225 |
0.618 |
| 195 |
247351_at |
AT5G63790
|
ANAC102, NAC domain containing protein 102, NAC102, NAC domain containing protein 102 |
-0.224 |
0.101 |
| 196 |
248969_at |
AT5G45310
|
unknown |
-0.219 |
-0.064 |
| 197 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
-0.218 |
0.223 |
| 198 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.218 |
0.691 |
| 199 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.217 |
0.704 |
| 200 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
-0.216 |
-0.057 |
| 201 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
-0.214 |
0.323 |
| 202 |
267077_at |
AT2G40970
|
MYBC1 |
-0.214 |
0.263 |
| 203 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
-0.212 |
0.069 |
| 204 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.209 |
0.617 |
| 205 |
263934_at |
AT2G35950
|
EDA12, embryo sac development arrest 12 |
-0.203 |
0.251 |
| 206 |
262580_at |
AT1G15330
|
[Cystathionine beta-synthase (CBS) protein] |
-0.202 |
0.203 |
| 207 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
-0.202 |
0.322 |
| 208 |
267150_at |
AT2G23510
|
SDT, spermidine disinapoyl acyltransferase |
-0.200 |
0.131 |
| 209 |
262803_at |
AT1G21000
|
[PLATZ transcription factor family protein] |
-0.199 |
0.216 |
| 210 |
245330_at |
AT4G14930
|
[Survival protein SurE-like phosphatase/nucleotidase] |
-0.196 |
0.187 |
| 211 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
-0.194 |
0.342 |
| 212 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
-0.194 |
0.700 |
| 213 |
263545_at |
AT2G21560
|
unknown |
-0.189 |
-0.180 |
| 214 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
-0.187 |
0.344 |
| 215 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
-0.187 |
0.534 |
| 216 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.187 |
0.037 |
| 217 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.185 |
0.267 |
| 218 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
-0.180 |
0.271 |
| 219 |
246523_at |
AT5G15850
|
ATCOL1, BBX2, B-box domain protein 2, COL1, CONSTANS-like 1 |
-0.179 |
0.465 |
| 220 |
248004_at |
AT5G56230
|
PRA1.G2, prenylated RAB acceptor 1.G2 |
-0.179 |
0.435 |
| 221 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.179 |
-0.077 |
| 222 |
263005_at |
AT1G54540
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.178 |
-0.199 |
| 223 |
247485_at |
AT5G62120
|
ARR23, response regulator 23, RR23, response regulator 23 |
-0.177 |
0.044 |
| 224 |
247474_at |
AT5G62280
|
unknown |
-0.177 |
0.145 |
| 225 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.177 |
0.307 |
| 226 |
259015_at |
AT3G07350
|
unknown |
-0.176 |
0.233 |
| 227 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
-0.175 |
0.268 |
| 228 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.174 |
0.368 |
| 229 |
264889_at |
AT1G23050
|
hydroxyproline-rich glycoprotein family protein |
-0.173 |
0.379 |
| 230 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
-0.173 |
0.299 |
| 231 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-0.170 |
-0.283 |
| 232 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.170 |
-0.005 |
| 233 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
-0.169 |
0.052 |
| 234 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
-0.168 |
0.023 |
| 235 |
248374_at |
AT5G51870
|
AGL71, AGAMOUS-like 71 |
-0.167 |
0.172 |
| 236 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
-0.167 |
0.019 |
| 237 |
256621_at |
AT3G24450
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.166 |
0.154 |
| 238 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.165 |
0.120 |
| 239 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.164 |
0.012 |
| 240 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.163 |
0.313 |
| 241 |
248276_at |
AT5G53550
|
ATYSL3, YELLOW STRIPE LIKE 3, YSL3, YELLOW STRIPE like 3 |
-0.161 |
0.023 |
| 242 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
-0.161 |
0.483 |
| 243 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.160 |
0.391 |
| 244 |
259418_at |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
-0.159 |
0.258 |
| 245 |
260199_at |
AT1G67590
|
[Remorin family protein] |
-0.159 |
-0.122 |
| 246 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
-0.157 |
0.335 |
| 247 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
-0.157 |
-0.008 |
| 248 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
-0.156 |
0.411 |
| 249 |
246279_at |
AT4G36740
|
ATHB40, homeobox protein 40, HB-5, HB40, homeobox protein 40 |
-0.154 |
-0.025 |
| 250 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
-0.153 |
0.154 |
| 251 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.153 |
0.113 |
| 252 |
265983_at |
AT2G18550
|
ATHB21, homeobox protein 21, HB-2, homeobox-2, HB21, homeobox protein 21 |
-0.153 |
0.146 |
| 253 |
258262_at |
AT3G15770
|
unknown |
-0.152 |
0.367 |
| 254 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
-0.152 |
-0.084 |
| 255 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.151 |
0.196 |
| 256 |
265263_at |
AT2G42940
|
AHL16, AT-hook motif nuclear-localized protein 16, TEK, TRANSPOSABLE ELEMENT SILENCING VIA AT-HOOK |
-0.151 |
0.143 |
| 257 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
-0.149 |
0.152 |
| 258 |
266363_at |
AT2G41250
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.149 |
0.422 |
| 259 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
-0.147 |
0.404 |
| 260 |
245098_at |
AT2G40940
|
ERS, ETHYLENE RESPONSE SENSOR, ERS1, ethylene response sensor 1 |
-0.147 |
0.089 |
| 261 |
250028_at |
AT5G18130
|
unknown |
-0.146 |
0.036 |
| 262 |
259596_at |
AT1G28130
|
GH3.17 |
-0.144 |
0.208 |
| 263 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
-0.144 |
0.442 |
| 264 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
-0.143 |
0.083 |
| 265 |
264338_at |
AT1G70300
|
KUP6, K+ uptake permease 6 |
-0.143 |
-0.148 |
| 266 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.142 |
0.093 |
| 267 |
254919_at |
AT4G11360
|
RHA1B, RING-H2 finger A1B |
-0.142 |
0.145 |
| 268 |
260010_at |
AT1G68020
|
ATTPS6, TPS6, TREHALOSE -6-PHOSPHATASE SYNTHASE S6 |
-0.141 |
0.077 |
| 269 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.141 |
0.196 |
| 270 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
-0.141 |
0.375 |
| 271 |
264815_at |
AT1G03620
|
[ELMO/CED-12 family protein] |
-0.141 |
0.027 |
| 272 |
256290_at |
AT3G12203
|
scpl17, serine carboxypeptidase-like 17 |
-0.140 |
0.072 |
| 273 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.140 |
-0.349 |
| 274 |
259901_at |
AT1G74180
|
AtRLP14, receptor like protein 14, RLP14, receptor like protein 14 |
-0.139 |
0.021 |
| 275 |
267393_at |
AT2G44500
|
[O-fucosyltransferase family protein] |
-0.138 |
0.150 |
| 276 |
246582_at |
AT1G31750
|
[proline-rich family protein] |
-0.138 |
0.083 |
| 277 |
260983_at |
AT1G53560
|
[Ribosomal protein L18ae family] |
-0.138 |
0.351 |
| 278 |
247835_at |
AT5G57910
|
unknown |
-0.138 |
0.152 |
| 279 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
-0.138 |
0.422 |
| 280 |
248487_at |
AT5G51070
|
CLPD, ERD1, EARLY RESPONSIVE TO DEHYDRATION 1, SAG15, SENESCENCE ASSOCIATED GENE 15 |
-0.137 |
0.209 |
| 281 |
249869_at |
AT5G23050
|
AAE17, acyl-activating enzyme 17 |
-0.137 |
0.085 |
| 282 |
253994_at |
AT4G26080
|
ABI1, ABA INSENSITIVE 1, AtABI1 |
-0.137 |
0.174 |
| 283 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
-0.137 |
0.393 |
| 284 |
254688_at |
AT4G13830
|
DJC26, DNA J protein C26, J20, DNAJ-like 20 |
-0.136 |
0.195 |
| 285 |
250467_at |
AT5G10100
|
TPPI, trehalose-6-phosphate phosphatase I |
-0.136 |
0.116 |
| 286 |
261026_at |
AT1G01240
|
unknown |
-0.136 |
-0.018 |
| 287 |
246595_at |
AT5G14780
|
FDH, formate dehydrogenase |
-0.136 |
0.255 |
| 288 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.135 |
0.206 |
| 289 |
260144_at |
AT1G71960
|
ABCG25, ATP-binding casette G25, ATABCG25, Arabidopsis thaliana ATP-binding cassette G25 |
-0.135 |
0.305 |
| 290 |
259340_at |
AT3G03870
|
unknown |
-0.135 |
0.171 |
| 291 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.135 |
0.239 |
| 292 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
-0.135 |
0.440 |
| 293 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-0.135 |
-0.284 |
| 294 |
259976_at |
AT1G76560
|
CP12-3, CP12 domain-containing protein 3 |
-0.134 |
0.378 |
| 295 |
246951_at |
AT5G04880
|
[expressed protein] |
-0.134 |
-0.029 |
| 296 |
247819_at |
AT5G58350
|
WNK4, with no lysine (K) kinase 4, ZIK2 |
-0.134 |
0.376 |
| 297 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.133 |
0.253 |
| 298 |
256181_at |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
-0.133 |
0.090 |
| 299 |
246996_at |
AT5G67420
|
ASL39, ASYMMETRIC LEAVES2-LIKE 39, LBD37, LOB domain-containing protein 37 |
-0.131 |
0.085 |
| 300 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
-0.130 |
-0.032 |
| 301 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-0.130 |
0.543 |
| 302 |
253812_at |
AT4G28240
|
[Wound-responsive family protein] |
-0.130 |
0.244 |
| 303 |
257764_at |
AT3G23010
|
AtRLP36, receptor like protein 36, RLP36, receptor like protein 36 |
-0.128 |
0.160 |
| 304 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.128 |
0.330 |
| 305 |
248455_at |
AT5G51360
|
[Transcription elongation factor (TFIIS) family protein] |
-0.128 |
0.091 |
| 306 |
248040_at |
AT5G55970
|
[RING/U-box superfamily protein] |
-0.128 |
0.349 |
| 307 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.128 |
-0.003 |
| 308 |
249937_at |
AT5G22470
|
[NAD+ ADP-ribosyltransferases] |
-0.128 |
0.043 |
| 309 |
265266_at |
AT2G42890
|
AML2, MEI2-like 2, ML2, MEI2-like 2 |
-0.127 |
0.154 |
| 310 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
-0.127 |
0.043 |
| 311 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
-0.126 |
0.097 |
| 312 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.125 |
0.309 |
| 313 |
253305_at |
AT4G33666
|
unknown |
-0.125 |
0.243 |
| 314 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
-0.125 |
0.247 |
| 315 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
-0.124 |
0.696 |