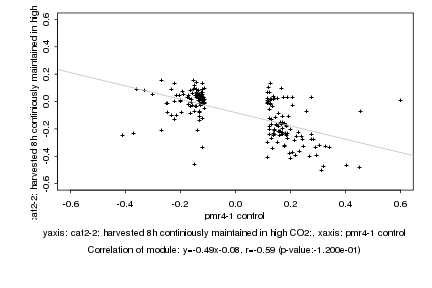
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
pmr4-1 control |
Log2 signal ratio
cat2-2; harvested 8h continiously maintained in high CO2; |
| 1 |
248918_at |
AT5G45890
|
AtSAG12, SAG12, senescence-associated gene 12 |
0.599 |
0.008 |
| 2 |
261684_at |
AT1G47400
|
unknown |
0.454 |
-0.066 |
| 3 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.451 |
-0.477 |
| 4 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.402 |
-0.465 |
| 5 |
259385_at |
AT1G13470
|
unknown |
0.339 |
-0.334 |
| 6 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.324 |
-0.324 |
| 7 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.317 |
-0.471 |
| 8 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.312 |
-0.502 |
| 9 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.303 |
-0.314 |
| 10 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.294 |
-0.388 |
| 11 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.288 |
-0.333 |
| 12 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
0.283 |
-0.272 |
| 13 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.276 |
-0.273 |
| 14 |
267345_at |
AT2G44240
|
unknown |
0.276 |
0.036 |
| 15 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.275 |
-0.237 |
| 16 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.267 |
-0.399 |
| 17 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
0.257 |
-0.071 |
| 18 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
0.245 |
-0.328 |
| 19 |
264580_at |
AT1G05340
|
unknown |
0.242 |
-0.277 |
| 20 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.237 |
-0.251 |
| 21 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.231 |
-0.364 |
| 22 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
0.227 |
-0.221 |
| 23 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.220 |
-0.250 |
| 24 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
0.215 |
-0.390 |
| 25 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.212 |
-0.278 |
| 26 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.206 |
-0.368 |
| 27 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.205 |
-0.022 |
| 28 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
0.204 |
0.034 |
| 29 |
249454_at |
AT5G39520
|
unknown |
0.197 |
-0.411 |
| 30 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
0.197 |
-0.203 |
| 31 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.195 |
-0.375 |
| 32 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.191 |
-0.105 |
| 33 |
252427_at |
AT3G47640
|
PYE, POPEYE |
0.189 |
0.033 |
| 34 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.187 |
-0.266 |
| 35 |
264958_at |
AT1G76960
|
unknown |
0.186 |
-0.233 |
| 36 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
0.184 |
-0.178 |
| 37 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.183 |
-0.174 |
| 38 |
249377_at |
AT5G40690
|
unknown |
0.183 |
-0.243 |
| 39 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.182 |
-0.230 |
| 40 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.177 |
-0.324 |
| 41 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.176 |
-0.157 |
| 42 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.175 |
-0.320 |
| 43 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.173 |
0.031 |
| 44 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.171 |
-0.225 |
| 45 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.170 |
-0.107 |
| 46 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
0.170 |
-0.236 |
| 47 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.168 |
-0.148 |
| 48 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.168 |
-0.192 |
| 49 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.166 |
0.099 |
| 50 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.164 |
-0.165 |
| 51 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.164 |
-0.239 |
| 52 |
263851_at |
AT2G04460
|
unknown |
0.161 |
-0.156 |
| 53 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.161 |
-0.217 |
| 54 |
248092_at |
AT5G55170
|
ATSUMO3, SUM3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO 3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO3, small ubiquitin-like modifier 3 |
0.161 |
-0.147 |
| 55 |
266464_at |
AT2G47800
|
ABCC4, ATP-binding cassette C4, ATMRP4, multidrug resistance-associated protein 4, EST3, MRP4, multidrug resistance-associated protein 4 |
0.158 |
-0.215 |
| 56 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.157 |
-0.242 |
| 57 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.156 |
-0.214 |
| 58 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.155 |
-0.084 |
| 59 |
245611_at |
AT4G14390
|
[Ankyrin repeat family protein] |
0.155 |
-0.181 |
| 60 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
0.152 |
-0.299 |
| 61 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.150 |
0.024 |
| 62 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.148 |
-0.170 |
| 63 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.147 |
-0.165 |
| 64 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.144 |
-0.111 |
| 65 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
0.142 |
-0.234 |
| 66 |
259065_at |
AT3G07520
|
ATGLR1.4, GLR1.4, glutamate receptor 1.4 |
0.141 |
-0.198 |
| 67 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.140 |
-0.208 |
| 68 |
246236_at |
AT4G36470
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.139 |
0.025 |
| 69 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.138 |
-0.248 |
| 70 |
248814_at |
AT5G46910
|
[Transcription factor jumonji (jmj) family protein / zinc finger (C5HC2 type) family protein] |
0.137 |
0.040 |
| 71 |
262085_at |
AT1G56060
|
unknown |
0.136 |
-0.227 |
| 72 |
260460_at |
AT1G68230
|
[Reticulon family protein] |
0.135 |
0.017 |
| 73 |
255912_at |
AT1G66960
|
LUP5 |
0.134 |
-0.337 |
| 74 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
0.133 |
-0.035 |
| 75 |
263861_at |
AT2G04560
|
AtLpxB, LpxB, lipid X B |
0.131 |
0.018 |
| 76 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.130 |
-0.131 |
| 77 |
248971_at |
AT5G45000
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.130 |
-0.267 |
| 78 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.129 |
-0.166 |
| 79 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.127 |
0.133 |
| 80 |
248709_at |
AT5G48470
|
PRDA1, PEP-Related Development Arrested 1 |
0.126 |
-0.008 |
| 81 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
0.125 |
-0.020 |
| 82 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
0.124 |
0.020 |
| 83 |
262507_at |
AT1G11330
|
[S-locus lectin protein kinase family protein] |
0.123 |
-0.058 |
| 84 |
249462_at |
AT5G39680
|
EMB2744, EMBRYO DEFECTIVE 2744 |
0.123 |
0.072 |
| 85 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.122 |
-0.199 |
| 86 |
267412_at |
AT2G34940
|
BP80-3;2, binding protein of 80 kDa 3;2, VSR3;2, VACUOLAR SORTING RECEPTOR 3;2, VSR5, VACUOLAR SORTING RECEPTOR 5 |
0.121 |
-0.237 |
| 87 |
256050_at |
AT1G07000
|
ATEXO70B2, exocyst subunit exo70 family protein B2, EXO70B2, exocyst subunit exo70 family protein B2 |
0.121 |
-0.123 |
| 88 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
0.121 |
-0.182 |
| 89 |
266738_at |
AT2G47010
|
unknown |
0.120 |
0.106 |
| 90 |
251113_at |
AT5G01370
|
ACI1, ALC-interacting protein 1, TRM29, TON1 Recruiting Motif 29 |
0.117 |
0.002 |
| 91 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.116 |
-0.405 |
| 92 |
266309_at |
AT2G27140
|
[HSP20-like chaperones superfamily protein] |
0.115 |
0.027 |
| 93 |
254153_at |
AT4G24450
|
ATGWD2, GWD3, PWD, phosphoglucan, water dikinase |
0.115 |
0.066 |
| 94 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
0.115 |
0.009 |
| 95 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
0.114 |
-0.004 |
| 96 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.113 |
-0.293 |
| 97 |
260728_at |
AT1G48210
|
[Protein kinase superfamily protein] |
0.113 |
-0.008 |
| 98 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
-0.413 |
-0.247 |
| 99 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
-0.374 |
-0.228 |
| 100 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.362 |
0.091 |
| 101 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
-0.333 |
0.083 |
| 102 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
-0.304 |
0.054 |
| 103 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.272 |
0.157 |
| 104 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.271 |
-0.211 |
| 105 |
251491_at |
AT3G59480
|
[pfkB-like carbohydrate kinase family protein] |
-0.251 |
-0.011 |
| 106 |
249726_at |
AT5G35480
|
unknown |
-0.249 |
-0.074 |
| 107 |
259511_at |
AT1G12520
|
ATCCS, copper chaperone for SOD1, CCS, copper chaperone for SOD1 |
-0.248 |
-0.013 |
| 108 |
247800_at |
AT5G58570
|
unknown |
-0.235 |
-0.098 |
| 109 |
252520_at |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
-0.235 |
0.092 |
| 110 |
256096_at |
AT1G13650
|
unknown |
-0.224 |
0.132 |
| 111 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.224 |
-0.125 |
| 112 |
266165_at |
AT2G28190
|
CSD2, copper/zinc superoxide dismutase 2, CZSOD2, COPPER/ZINC SUPEROXIDE DISMUTASE 2 |
-0.222 |
0.007 |
| 113 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.218 |
-0.099 |
| 114 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.217 |
0.047 |
| 115 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.207 |
0.045 |
| 116 |
254193_at |
AT4G23870
|
unknown |
-0.203 |
0.004 |
| 117 |
255378_at |
AT4G03550
|
ATGSL05, glucan synthase-like 5, ATGSL5, EED3, enhancer of edr1 3, GSL05, GSL05, glucan synthase-like 5, GSL5, GLUCAN SYNTHASE-LIKE 5, PMR4, POWDERY MILDEW RESISTANT 4 |
-0.200 |
0.008 |
| 118 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.198 |
-0.073 |
| 119 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
-0.193 |
0.080 |
| 120 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.191 |
0.054 |
| 121 |
267038_at |
AT2G38480
|
[Uncharacterised protein family (UPF0497)] |
-0.177 |
0.036 |
| 122 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.172 |
0.048 |
| 123 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
-0.171 |
-0.015 |
| 124 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.168 |
0.024 |
| 125 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.166 |
-0.033 |
| 126 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
-0.165 |
-0.081 |
| 127 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.165 |
0.085 |
| 128 |
246770_at |
AT5G27460
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.163 |
-0.002 |
| 129 |
251109_at |
AT5G01600
|
ATFER1, ARABIDOPSIS THALIANA FERRETIN 1, FER1, ferretin 1 |
-0.162 |
0.018 |
| 130 |
250327_at |
AT5G12050
|
unknown |
-0.158 |
0.059 |
| 131 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
-0.158 |
-0.025 |
| 132 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
-0.153 |
0.094 |
| 133 |
247474_at |
AT5G62280
|
unknown |
-0.153 |
0.158 |
| 134 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.152 |
0.119 |
| 135 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
-0.152 |
-0.456 |
| 136 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
-0.150 |
0.102 |
| 137 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
-0.149 |
-0.067 |
| 138 |
264809_at |
AT1G08830
|
CSD1, copper/zinc superoxide dismutase 1 |
-0.148 |
-0.032 |
| 139 |
259012_at |
AT3G07360
|
ATPUB9, ARABIDOPSIS THALIANA PLANT U-BOX 9, PUB9, plant U-box 9 |
-0.147 |
0.049 |
| 140 |
245657_at |
AT1G56720
|
[Protein kinase superfamily protein] |
-0.145 |
0.072 |
| 141 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
-0.144 |
-0.030 |
| 142 |
258150_at |
AT3G18160
|
PEX3-1, peroxin 3-1 |
-0.143 |
0.033 |
| 143 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.142 |
0.094 |
| 144 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.142 |
0.140 |
| 145 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
-0.140 |
-0.206 |
| 146 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.140 |
0.058 |
| 147 |
250901_at |
AT5G03530
|
ATRAB, ATRAB ALPHA, ATRAB18B, ATRABC2A, ARABIDOPSIS THALIANA RAB GTPASE HOMOLOG C2A, RABC2A, RAB GTPase homolog C2A |
-0.140 |
0.056 |
| 148 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.139 |
0.060 |
| 149 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
-0.138 |
0.082 |
| 150 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.137 |
0.031 |
| 151 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.137 |
0.011 |
| 152 |
264684_at |
AT1G65590
|
ATHEX1, HEXO3, beta-hexosaminidase 3 |
-0.137 |
0.025 |
| 153 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-0.136 |
0.038 |
| 154 |
259766_at |
AT1G64360
|
unknown |
-0.133 |
-0.109 |
| 155 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
-0.133 |
-0.132 |
| 156 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.132 |
0.009 |
| 157 |
254683_at |
AT4G13800
|
unknown |
-0.132 |
0.041 |
| 158 |
259570_at |
AT1G20440
|
AtCOR47, COR47, cold-regulated 47, RD17 |
-0.131 |
0.036 |
| 159 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
-0.131 |
-0.076 |
| 160 |
257925_at |
AT3G23170
|
unknown |
-0.131 |
-0.008 |
| 161 |
247597_at |
AT5G60860
|
AtRABA1f, RAB GTPase homolog A1F, RABA1f, RAB GTPase homolog A1F |
-0.131 |
0.052 |
| 162 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.131 |
0.023 |
| 163 |
258262_at |
AT3G15770
|
unknown |
-0.131 |
-0.071 |
| 164 |
267096_at |
AT2G38180
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.129 |
-0.025 |
| 165 |
258189_at |
AT3G17860
|
JAI3, JASMONATE-INSENSITIVE 3, JAZ3, jasmonate-zim-domain protein 3, TIFY6B |
-0.129 |
0.049 |
| 166 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.128 |
-0.010 |
| 167 |
247903_at |
AT5G57340
|
unknown |
-0.127 |
-0.011 |
| 168 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.125 |
0.058 |
| 169 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
-0.124 |
0.072 |
| 170 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-0.124 |
0.018 |
| 171 |
250694_at |
AT5G06710
|
HAT14, homeobox from Arabidopsis thaliana |
-0.124 |
0.037 |
| 172 |
260129_at |
AT1G36380
|
unknown |
-0.124 |
-0.020 |
| 173 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
-0.122 |
0.089 |
| 174 |
248911_at |
AT5G45830
|
ATDOG1, DOG1, DELAY OF GERMINATION 1, GAAS5, germination ability after storage 5, GSQ5, GLUCOSE SENSING QTL 5 |
-0.122 |
0.040 |
| 175 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
-0.122 |
-0.334 |
| 176 |
266406_at |
AT2G38570
|
unknown |
-0.122 |
-0.018 |
| 177 |
257151_at |
AT3G27200
|
[Cupredoxin superfamily protein] |
-0.122 |
0.069 |
| 178 |
246462_at |
AT5G16940
|
[carbon-sulfur lyases] |
-0.121 |
-0.000 |
| 179 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.121 |
0.133 |
| 180 |
263931_at |
AT2G36220
|
unknown |
-0.121 |
0.056 |
| 181 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.121 |
0.032 |
| 182 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.121 |
0.095 |
| 183 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
-0.120 |
-0.122 |
| 184 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.119 |
0.010 |
| 185 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.119 |
-0.002 |
| 186 |
251665_at |
AT3G57040
|
ARR9, response regulator 9, ATRR4, RESPONSE REGULATOR 4 |
-0.118 |
0.031 |
| 187 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
-0.118 |
0.032 |
| 188 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.117 |
0.017 |
| 189 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.116 |
0.101 |
| 190 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-0.116 |
-0.050 |
| 191 |
247726_at |
AT5G59430
|
ATTRP1, TELOMERE REPEAT BINDING PROTEIN 1, TRP1, telomeric repeat binding protein 1 |
-0.116 |
-0.005 |
| 192 |
251816_at |
AT3G55005
|
TON1B, tonneau 1b |
-0.115 |
0.016 |
| 193 |
265116_at |
AT1G62480
|
[Vacuolar calcium-binding protein-related] |
-0.114 |
-0.012 |