|
probeID |
AGICode |
Annotation |
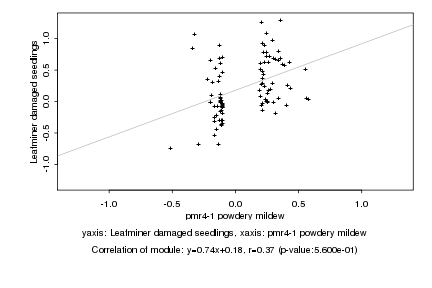
Log2 signal ratio
pmr4-1 powdery mildew |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
248918_at |
AT5G45890
|
AtSAG12, SAG12, senescence-associated gene 12 |
0.576 |
0.044 |
| 2 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.560 |
0.062 |
| 3 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
0.553 |
0.514 |
| 4 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.429 |
0.215 |
| 5 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.423 |
0.636 |
| 6 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.406 |
0.257 |
| 7 |
267345_at |
AT2G44240
|
unknown |
0.397 |
-0.061 |
| 8 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.386 |
0.580 |
| 9 |
249454_at |
AT5G39520
|
unknown |
0.367 |
0.603 |
| 10 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.356 |
1.304 |
| 11 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.351 |
0.689 |
| 12 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.340 |
0.797 |
| 13 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.335 |
0.653 |
| 14 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
0.334 |
0.060 |
| 15 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
0.315 |
-0.177 |
| 16 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.311 |
0.675 |
| 17 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.297 |
-0.002 |
| 18 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.297 |
0.685 |
| 19 |
264580_at |
AT1G05340
|
unknown |
0.287 |
0.981 |
| 20 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
0.286 |
0.298 |
| 21 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.273 |
0.207 |
| 22 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
0.269 |
0.721 |
| 23 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
0.258 |
0.182 |
| 24 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.256 |
0.627 |
| 25 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.252 |
0.141 |
| 26 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
0.249 |
0.012 |
| 27 |
251419_at |
AT3G60470
|
unknown |
0.246 |
-0.009 |
| 28 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.244 |
0.011 |
| 29 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.242 |
0.783 |
| 30 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.242 |
0.731 |
| 31 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.240 |
1.094 |
| 32 |
255912_at |
AT1G66960
|
LUP5 |
0.237 |
0.037 |
| 33 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.226 |
0.241 |
| 34 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.226 |
0.621 |
| 35 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.224 |
0.902 |
| 36 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.220 |
0.442 |
| 37 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.215 |
0.795 |
| 38 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.214 |
-0.137 |
| 39 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.214 |
-0.016 |
| 40 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.212 |
0.935 |
| 41 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.212 |
0.371 |
| 42 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.210 |
0.489 |
| 43 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
0.207 |
0.289 |
| 44 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.205 |
1.262 |
| 45 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
0.203 |
0.277 |
| 46 |
259012_at |
AT3G07360
|
ATPUB9, ARABIDOPSIS THALIANA PLANT U-BOX 9, PUB9, plant U-box 9 |
0.201 |
-0.054 |
| 47 |
266376_at |
AT2G14620
|
XTH10, xyloglucan endotransglucosylase/hydrolase 10 |
0.197 |
0.093 |
| 48 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.195 |
0.518 |
| 49 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.195 |
0.609 |
| 50 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.189 |
0.183 |
| 51 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.516 |
-0.738 |
| 52 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-0.342 |
0.858 |
| 53 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
-0.327 |
1.075 |
| 54 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.296 |
-0.673 |
| 55 |
252520_at |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
-0.225 |
0.362 |
| 56 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.200 |
0.668 |
| 57 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.199 |
-0.005 |
| 58 |
255378_at |
AT4G03550
|
ATGSL05, glucan synthase-like 5, ATGSL5, EED3, enhancer of edr1 3, GSL05, GSL05, glucan synthase-like 5, GSL5, GLUCAN SYNTHASE-LIKE 5, PMR4, POWDERY MILDEW RESISTANT 4 |
-0.198 |
-0.006 |
| 59 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
-0.193 |
0.101 |
| 60 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-0.185 |
0.315 |
| 61 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.171 |
-0.526 |
| 62 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.171 |
-0.307 |
| 63 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.170 |
-0.240 |
| 64 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
-0.168 |
-0.067 |
| 65 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.160 |
0.540 |
| 66 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.152 |
-0.212 |
| 67 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
-0.151 |
-0.435 |
| 68 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.145 |
-0.065 |
| 69 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.142 |
-0.678 |
| 70 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
-0.138 |
0.320 |
| 71 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.134 |
0.402 |
| 72 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.131 |
0.699 |
| 73 |
248853_at |
AT5G46570
|
BSK2, brassinosteroid-signaling kinase 2 |
-0.130 |
-0.300 |
| 74 |
265418_at |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
-0.128 |
0.897 |
| 75 |
266066_at |
AT2G18800
|
ATXTH21, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 21, XTH21, xyloglucan endotransglucosylase/hydrolase 21 |
-0.126 |
0.004 |
| 76 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
-0.126 |
0.613 |
| 77 |
259643_at |
AT1G68890
|
PHYLLO, PHYLLO |
-0.124 |
-0.147 |
| 78 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.123 |
0.115 |
| 79 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
-0.122 |
0.054 |
| 80 |
254281_at |
AT4G22840
|
[Sodium Bile acid symporter family] |
-0.122 |
0.078 |
| 81 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.122 |
-0.015 |
| 82 |
266543_at |
AT2G35075
|
unknown |
-0.121 |
0.030 |
| 83 |
257034_at |
AT3G19184
|
[AP2/B3-like transcriptional factor family protein] |
-0.116 |
-0.064 |
| 84 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.116 |
-0.365 |
| 85 |
267089_at |
AT2G38300
|
[myb-like HTH transcriptional regulator family protein] |
-0.113 |
-0.374 |
| 86 |
253148_at |
AT4G35620
|
CYCB2;2, Cyclin B2;2 |
-0.112 |
0.028 |
| 87 |
259346_at |
AT3G03910
|
glutamate dehydrogenase 3 |
-0.112 |
-0.287 |
| 88 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
-0.111 |
-0.142 |
| 89 |
264050_at |
AT2G22350
|
[similar to reverse transcriptase-related [Arabidopsis thaliana] (TAIR:AT4G12275.1)] |
-0.110 |
-0.035 |
| 90 |
251058_at |
AT5G01790
|
unknown |
-0.109 |
-0.294 |
| 91 |
261248_at |
AT1G20030
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.109 |
0.709 |
| 92 |
257971_at |
AT3G27530
|
GC6, golgin candidate 6, MAG4, MAIGO 4 |
-0.108 |
-0.083 |
| 93 |
266337_at |
AT2G32390
|
ATGLR3.5, glutamate receptor 3.5, GLR3.5, glutamate receptor 3.5, GLR6 |
-0.108 |
-0.016 |
| 94 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.108 |
0.474 |
| 95 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.107 |
-0.348 |
| 96 |
256978_at |
AT3G21110
|
ATPURC, PUR7, purin 7, PURC, PURIN C |
-0.106 |
-0.189 |
| 97 |
261514_at |
AT1G71870
|
[MATE efflux family protein] |
-0.106 |
-0.090 |
| 98 |
262007_at |
AT1G64580
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.104 |
-0.062 |
| 99 |
258701_at |
AT3G09720
|
AtRH57, RH57, RNA helicase 57 |
-0.103 |
-0.079 |