|
probeID |
AGICode |
Annotation |
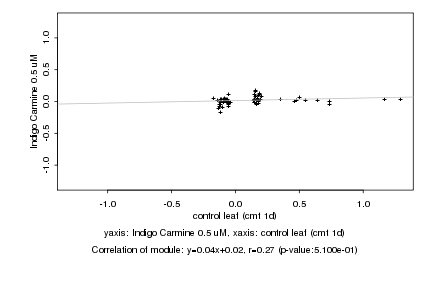
Log2 signal ratio
control leaf (cmt 1d) |
Log2 signal ratio
Indigo Carmine 0.5 uM |
| 1 |
248949_at |
AT5G45570
|
[Ulp1 protease family protein] |
1.289 |
0.046 |
| 2 |
246888_at |
AT5G26270
|
unknown |
1.163 |
0.036 |
| 3 |
251458_at |
AT3G60170
|
[copia-like retrotransposon family, has a 7.6e-229 P-value blast match to gb|AAO73527.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
0.734 |
0.012 |
| 4 |
246869_at |
AT5G26020
|
[similar to Glutamic acid-rich protein precursor (GB:P13816)] |
0.731 |
-0.045 |
| 5 |
263367_at |
AT2G20460
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
0.638 |
0.018 |
| 6 |
263346_at |
AT2G05660
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 0. P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
0.545 |
0.026 |
| 7 |
266807_at |
AT2G29920
|
unknown |
0.500 |
0.072 |
| 8 |
265503_at |
AT2G15510
|
[non-LTR retrotransposon family (LINE), has a 3.0e-20 P-value blast match to GB:NP_038602 L1 repeat, Tf subfamily, member 18 (LINE-element) (Mus musculus)] |
0.472 |
0.024 |
| 9 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.457 |
0.002 |
| 10 |
250979_at |
AT5G03090
|
unknown |
0.346 |
0.032 |
| 11 |
259584_at |
AT1G28080
|
[RING finger protein] |
0.198 |
0.088 |
| 12 |
260557_at |
AT2G43610
|
[Chitinase family protein] |
0.194 |
0.034 |
| 13 |
245546_at |
AT4G15290
|
ATCSLB05, ATCSLB5, CELLULOSE SYNTHASE LIKE 5, CSLB05 |
0.187 |
0.141 |
| 14 |
251843_x_at |
AT3G54590
|
ATHRGP1, hydroxyproline-rich glycoprotein, HRGP1, hydroxyproline-rich glycoprotein |
0.183 |
0.114 |
| 15 |
258354_at |
AT3G14320
|
[Zinc finger, C3HC4 type (RING finger) family protein] |
0.175 |
0.028 |
| 16 |
264358_at |
AT1G03180
|
unknown |
0.175 |
-0.031 |
| 17 |
250770_at |
AT5G05390
|
LAC12, laccase 12 |
0.175 |
0.098 |
| 18 |
260300_at |
AT1G80340
|
ATGA3OX2, ARABIDOPSIS THALIANA GIBBERELLIN-3-OXIDASE 2, GA3OX2, gibberellin 3-oxidase 2, GA4H |
0.174 |
0.001 |
| 19 |
248113_at |
AT5G55360
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
0.170 |
0.061 |
| 20 |
258199_at |
AT3G14030
|
[F-box associated ubiquitination effector family protein] |
0.160 |
-0.026 |
| 21 |
250136_at |
AT5G15380
|
DRM1, domains rearranged methylase 1 |
0.160 |
-0.041 |
| 22 |
254237_at |
AT4G23520
|
[Cysteine proteinases superfamily protein] |
0.158 |
0.029 |
| 23 |
250171_at |
AT5G14300
|
ATPHB5, prohibitin 5, PHB5, prohibitin 5 |
0.152 |
0.172 |
| 24 |
255761_at |
AT1G16770
|
unknown |
0.151 |
0.090 |
| 25 |
248926_at |
AT5G45880
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.151 |
0.176 |
| 26 |
257135_at |
AT3G12900
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.148 |
0.037 |
| 27 |
258240_at |
AT3G27660
|
OLE3, OLEOSIN 3, OLEO4, oleosin 4 |
0.145 |
0.115 |
| 28 |
258267_at |
AT3G15870
|
[Fatty acid desaturase family protein] |
0.145 |
0.028 |
| 29 |
250251_at |
AT5G13670
|
UMAMIT15, Usually multiple acids move in and out Transporters 15 |
0.142 |
-0.001 |
| 30 |
252301_at |
AT3G49162
|
|
0.142 |
-0.009 |
| 31 |
254009_at |
AT4G26390
|
[Pyruvate kinase family protein] |
0.142 |
0.033 |
| 32 |
261473_at |
AT1G14490
|
AHL28, AT-hook motif nuclear localized protein 28 |
0.141 |
0.022 |
| 33 |
245579_at |
AT4G14810
|
unknown |
-0.173 |
0.053 |
| 34 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
-0.145 |
0.024 |
| 35 |
247491_at |
AT5G61880
|
[Protein Transporter, Pam16] |
-0.136 |
-0.098 |
| 36 |
263515_at |
AT2G21640
|
unknown |
-0.132 |
-0.033 |
| 37 |
256248_at |
AT3G66652
|
[fip1 motif-containing protein] |
-0.129 |
-0.071 |
| 38 |
249034_at |
AT5G44160
|
AtIDD8, IDD8, INDETERMINATE DOMAIN 8, NUC, nutcracker |
-0.122 |
0.032 |
| 39 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
-0.119 |
-0.169 |
| 40 |
246409_at |
AT1G57700
|
[Protein kinase superfamily protein] |
-0.118 |
0.007 |
| 41 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
-0.118 |
-0.015 |
| 42 |
264753_at |
AT1G61490
|
[S-locus lectin protein kinase family protein] |
-0.117 |
-0.033 |
| 43 |
246325_at |
AT1G16540
|
ABA3, ABA DEFICIENT 3, ACI2, ALTERED CHLOROPLAST IMPORT 2, ATABA3, AtLOS5, LOS5, LOW OSMOTIC STRESS 5, SIR3, SIRTINOL RESISTANT 3 |
-0.114 |
0.034 |
| 44 |
266794_at |
AT2G02980
|
OTP85, ORGANELLE TRANSCRIPT PROCESSING 85 |
-0.102 |
-0.087 |
| 45 |
259877_at |
AT1G76710
|
ASHH1, ASH1 RELATED PROTEIN 1, ASHH1, ASH1-RELATED PROTEIN 1, SDG26, SET domain group 26 |
-0.098 |
0.035 |
| 46 |
245871_at |
AT1G26290
|
unknown |
-0.097 |
0.004 |
| 47 |
267467_at |
AT2G30580
|
BMI1A, DRIP2, DREB2A-interacting protein 2 |
-0.096 |
-0.001 |
| 48 |
263032_at |
AT1G23850
|
unknown |
-0.089 |
0.049 |
| 49 |
256311_at |
AT1G30330
|
ARF6, auxin response factor 6 |
-0.087 |
0.038 |
| 50 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.084 |
0.004 |
| 51 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
-0.082 |
0.037 |
| 52 |
259774_at |
AT1G29520
|
[AWPM-19-like family protein] |
-0.074 |
0.037 |
| 53 |
250566_at |
AT5G08070
|
TCP17, TCP domain protein 17 |
-0.071 |
0.030 |
| 54 |
247239_at |
AT5G64640
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.067 |
-0.013 |
| 55 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.067 |
0.044 |
| 56 |
253089_at |
AT4G36290
|
AtMORC1, CRT1, compromised recognition of TCV 1, MORC1, Microchidia 1 |
-0.062 |
-0.021 |
| 57 |
255169_x_at |
AT4G07940
|
unknown |
-0.060 |
0.111 |
| 58 |
264909_at |
AT2G17300
|
unknown |
-0.059 |
-0.011 |
| 59 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
-0.058 |
-0.033 |
| 60 |
260494_at |
AT2G41820
|
PXC3, PXY/TDR-correlated 3 |
-0.058 |
-0.016 |
| 61 |
255702_at |
AT4G00230
|
XSP1, xylem serine peptidase 1 |
-0.056 |
-0.063 |
| 62 |
249650_at |
AT5G37055
|
ATSWC6, SEF, SERRATED LEAVES AND EARLY FLOWERING |
-0.055 |
0.009 |
| 63 |
251967_at |
AT3G53390
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.041 |
-0.002 |