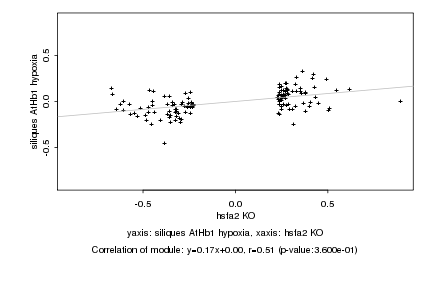
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
hsfa2 KO |
Log2 signal ratio
siliques AtHb1 hypoxia |
| 1 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
0.891 |
0.004 |
| 2 |
253532_at |
AT4G31570
|
unknown |
0.615 |
0.132 |
| 3 |
263285_at |
AT2G36120
|
DOT1, DEFECTIVELY ORGANIZED TRIBUTARIES 1 |
0.546 |
0.126 |
| 4 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
0.506 |
-0.069 |
| 5 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
0.503 |
-0.096 |
| 6 |
259686_at |
AT1G63100
|
[GRAS family transcription factor] |
0.488 |
0.242 |
| 7 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.447 |
-0.015 |
| 8 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.431 |
0.049 |
| 9 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.423 |
0.157 |
| 10 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
0.419 |
0.298 |
| 11 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.414 |
0.261 |
| 12 |
262974_at |
AT1G75550
|
[glycine-rich protein] |
0.403 |
-0.009 |
| 13 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
0.398 |
-0.045 |
| 14 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
0.379 |
0.099 |
| 15 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
0.378 |
0.095 |
| 16 |
258414_at |
AT3G17380
|
[TRAF-like family protein] |
0.376 |
-0.098 |
| 17 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
0.365 |
-0.019 |
| 18 |
261333_at |
AT1G44910
|
ATPRP40A, pre-mRNA-processing protein 40A, PRP40A, pre-mRNA-processing protein 40A |
0.361 |
0.336 |
| 19 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
0.354 |
0.088 |
| 20 |
254208_at |
AT4G24175
|
unknown |
0.351 |
0.152 |
| 21 |
252208_at |
AT3G50380
|
[INVOLVED IN: protein localization] |
0.349 |
0.113 |
| 22 |
258471_at |
AT3G06030
|
ANP3, NPK1-related protein kinase 3, AtANP3, MAPKKK12, NP3, NPK1-related protein kinase 3 |
0.328 |
0.109 |
| 23 |
264698_at |
AT1G70200
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.327 |
0.271 |
| 24 |
257724_at |
AT3G18510
|
unknown |
0.324 |
0.187 |
| 25 |
260230_at |
AT1G74500
|
ATBS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, bHLH135, basic helix-loop-helix protein 135, BS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, PRE3, PACLOBUTRAZOL RESISTANCE 3, TMO7, TARGET OF MONOPTEROS 7 |
0.322 |
-0.054 |
| 26 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
0.310 |
-0.249 |
| 27 |
258618_at |
AT3G02885
|
GASA5, GAST1 protein homolog 5 |
0.307 |
-0.080 |
| 28 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
0.306 |
0.110 |
| 29 |
249948_at |
AT5G19210
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.288 |
-0.086 |
| 30 |
266951_at |
AT2G18940
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.284 |
-0.023 |
| 31 |
248709_at |
AT5G48470
|
PRDA1, PEP-Related Development Arrested 1 |
0.282 |
0.086 |
| 32 |
262242_at |
AT1G48360
|
[zinc ion binding] |
0.281 |
0.136 |
| 33 |
247959_at |
AT5G57080
|
unknown |
0.281 |
0.097 |
| 34 |
248291_at |
AT5G53020
|
[Ribonuclease P protein subunit P38-related] |
0.281 |
0.120 |
| 35 |
263864_at |
AT2G04530
|
CPZ, TRZ2, TRNASE Z 2 |
0.276 |
-0.040 |
| 36 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
0.275 |
0.201 |
| 37 |
265522_at |
AT2G06210
|
ELF8, EARLY FLOWERING 8, VIP6, VERNALIZATION INDEPENDENCE 6 |
0.273 |
0.148 |
| 38 |
247425_at |
AT5G62550
|
unknown |
0.270 |
0.203 |
| 39 |
265831_at |
AT2G14460
|
unknown |
0.269 |
0.109 |
| 40 |
247235_at |
AT5G64580
|
EMB3144, EMBRYO DEFECTIVE 3144 |
0.268 |
0.072 |
| 41 |
258034_at |
AT3G21300
|
[RNA methyltransferase family protein] |
0.267 |
0.034 |
| 42 |
263535_at |
AT2G24970
|
unknown |
0.261 |
0.076 |
| 43 |
249852_at |
AT5G23270
|
ATSTP11, SUGAR TRANSPORTER 11, STP11, sugar transporter 11 |
0.259 |
-0.023 |
| 44 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
0.259 |
0.078 |
| 45 |
253078_at |
AT4G36180
|
[Leucine-rich receptor-like protein kinase family protein] |
0.257 |
0.123 |
| 46 |
256125_at |
AT1G18250
|
ATLP-1 |
0.252 |
0.055 |
| 47 |
247550_at |
AT5G61370
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.250 |
0.045 |
| 48 |
260204_at |
AT1G52900
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.248 |
-0.080 |
| 49 |
261521_at |
AT1G71830
|
ATSERK1, SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE 1, SERK1, somatic embryogenesis receptor-like kinase 1 |
0.248 |
0.166 |
| 50 |
245461_at |
AT4G17000
|
unknown |
0.247 |
0.130 |
| 51 |
252522_at |
AT3G46390
|
unknown |
0.247 |
0.021 |
| 52 |
245847_at |
AT5G11050
|
AtMYB64, myb domain protein 64, MYB64, myb domain protein 64, URP10, UAS-TAGGED ROOT PATTERNING10 |
0.247 |
-0.047 |
| 53 |
259290_at |
AT3G11520
|
CYC2, CYCLIN 2, CYCB1;3, CYCLIN B1;3 |
0.245 |
0.073 |
| 54 |
266618_at |
AT2G35480
|
unknown |
0.244 |
0.040 |
| 55 |
248139_at |
AT5G54970
|
unknown |
0.242 |
0.026 |
| 56 |
246746_at |
AT5G27820
|
[Ribosomal L18p/L5e family protein] |
0.238 |
0.011 |
| 57 |
248708_at |
AT5G48560
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.238 |
-0.137 |
| 58 |
264091_at |
AT1G79110
|
BRG2, BOI-related gene 2 |
0.237 |
-0.030 |
| 59 |
247594_at |
AT5G60800
|
[Heavy metal transport/detoxification superfamily protein ] |
0.237 |
0.190 |
| 60 |
251650_at |
AT3G57360
|
unknown |
0.237 |
0.101 |
| 61 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.234 |
0.081 |
| 62 |
265596_at |
AT2G20020
|
ATCAF1, CAF1 |
0.234 |
0.158 |
| 63 |
262693_at |
AT1G62780
|
unknown |
0.233 |
-0.025 |
| 64 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
0.232 |
0.022 |
| 65 |
255236_at |
AT4G05520
|
ATEHD2, EPS15 homology domain 2, EHD2, EPS15 homology domain 2 |
0.230 |
0.073 |
| 66 |
254662_at |
AT4G18270
|
ATTRANS11, ARABIDOPSIS THALIANA TRANSLOCASE 11, TRANS11, translocase 11 |
0.229 |
-0.121 |
| 67 |
254639_at |
AT4G18750
|
DOT4, DEFECTIVELY ORGANIZED TRIBUTARIES 4 |
0.227 |
0.045 |
| 68 |
247951_at |
AT5G57240
|
ORP4C, OSBP(oxysterol binding protein)-related protein 4C |
-0.673 |
0.152 |
| 69 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
-0.666 |
0.079 |
| 70 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
-0.645 |
-0.078 |
| 71 |
244932_at |
ATCG01060
|
PSAC |
-0.622 |
-0.024 |
| 72 |
250135_at |
AT5G15360
|
unknown |
-0.606 |
-0.089 |
| 73 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-0.606 |
0.010 |
| 74 |
250098_at |
AT5G17350
|
unknown |
-0.575 |
-0.025 |
| 75 |
245460_at |
AT4G16990
|
RLM3, RESISTANCE TO LEPTOSPHAERIA MACULANS 3 |
-0.569 |
-0.133 |
| 76 |
253284_at |
AT4G34150
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.549 |
-0.130 |
| 77 |
266019_at |
AT2G18750
|
[Calmodulin-binding protein] |
-0.534 |
-0.159 |
| 78 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
-0.514 |
-0.068 |
| 79 |
254169_at |
AT4G24290
|
[MAC/Perforin domain-containing protein] |
-0.492 |
-0.148 |
| 80 |
245290_at |
AT4G16490
|
[ARM repeat superfamily protein] |
-0.484 |
-0.204 |
| 81 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
-0.473 |
-0.055 |
| 82 |
254107_at |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
-0.471 |
-0.110 |
| 83 |
258282_at |
AT3G26910
|
[hydroxyproline-rich glycoprotein family protein] |
-0.466 |
0.129 |
| 84 |
251829_at |
AT3G55020
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
-0.456 |
-0.242 |
| 85 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
-0.453 |
-0.042 |
| 86 |
247474_at |
AT5G62280
|
unknown |
-0.449 |
0.007 |
| 87 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-0.448 |
0.117 |
| 88 |
256968_at |
AT3G21070
|
ATNADK-1, NAD KINASE 1, NADK1, NAD kinase 1 |
-0.440 |
-0.109 |
| 89 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
-0.408 |
-0.204 |
| 90 |
251842_at |
AT3G54580
|
[Proline-rich extensin-like family protein] |
-0.387 |
-0.448 |
| 91 |
262719_at |
AT1G43590
|
unknown |
-0.384 |
0.060 |
| 92 |
255015_at |
AT4G09980
|
EMB1691, EMBRYO DEFECTIVE 1691 |
-0.369 |
-0.138 |
| 93 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.369 |
-0.022 |
| 94 |
263614_at |
AT2G25240
|
AtCCP3, CCP3, conserved in ciliated species and in the land plants 3 |
-0.361 |
-0.107 |
| 95 |
253925_at |
AT4G26690
|
GDPDL3, Glycerophosphodiester phosphodiesterase (GDPD) like 3, GPDL2, GLYCEROPHOSPHODIESTERASE-LIKE 2, MRH5, MUTANT ROOT HAIR 5, SHV3, SHAVEN 3 |
-0.360 |
-0.167 |
| 96 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.357 |
0.060 |
| 97 |
263163_at |
AT1G03160
|
FZL, FZO-like |
-0.355 |
-0.144 |
| 98 |
250939_at |
AT5G03040
|
iqd2, IQ-domain 2 |
-0.355 |
-0.219 |
| 99 |
253437_at |
AT4G32460
|
unknown |
-0.345 |
-0.038 |
| 100 |
257690_at |
AT3G12830
|
SAUR72, SMALL AUXIN UPREGULATED 72 |
-0.344 |
-0.006 |
| 101 |
248756_at |
AT5G47560
|
ATSDAT, ATTDT, TONOPLAST DICARBOXYLATE TRANSPORTER, TDT, tonoplast dicarboxylate transporter |
-0.335 |
-0.026 |
| 102 |
260642_at |
AT1G53260
|
[LOCATED IN: endomembrane system] |
-0.327 |
-0.082 |
| 103 |
251187_at |
AT3G62770
|
AtATG18a, autophagy 18a, ATG18a, autophagy 18a, PEUP2, PEROXISOME UNUSUAL POSITIONING 2 |
-0.326 |
-0.109 |
| 104 |
258985_at |
AT3G08960
|
[ARM repeat superfamily protein] |
-0.325 |
-0.205 |
| 105 |
265823_at |
AT2G35760
|
[Uncharacterised protein family (UPF0497)] |
-0.324 |
-0.078 |
| 106 |
263806_at |
AT2G04305
|
[Magnesium transporter CorA-like family protein] |
-0.322 |
-0.162 |
| 107 |
251200_at |
AT3G63010
|
ATGID1B, GID1B, GA INSENSITIVE DWARF1B |
-0.320 |
-0.107 |
| 108 |
250231_at |
AT5G13910
|
LEP, LEAFY PETIOLE |
-0.309 |
-0.122 |
| 109 |
246284_at |
AT4G36780
|
BEH2, BES1/BZR1 homolog 2 |
-0.306 |
-0.184 |
| 110 |
267019_at |
AT2G39130
|
[Transmembrane amino acid transporter family protein] |
-0.302 |
-0.221 |
| 111 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.293 |
-0.024 |
| 112 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
-0.292 |
-0.190 |
| 113 |
267069_at |
AT2G41010
|
ATCAMBP25, calmodulin (CAM)-binding protein of 25 kDa, CAMBP25, calmodulin (CAM)-binding protein of 25 kDa |
-0.289 |
-0.008 |
| 114 |
249232_at |
AT5G42080
|
ADL1, dynamin-like protein, ADL1A, AG68, DL1, dynamin-like protein, DRP1A, DYNAMIN-RELATED PROTEIN 1A, RSW9, RADIAL SWELLING 9 |
-0.276 |
-0.044 |
| 115 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
-0.275 |
-0.109 |
| 116 |
246495_at |
AT5G16200
|
[50S ribosomal protein-related] |
-0.271 |
0.088 |
| 117 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.264 |
-0.063 |
| 118 |
257851_at |
AT3G12940
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.260 |
-0.045 |
| 119 |
261405_at |
AT1G18740
|
unknown |
-0.258 |
0.038 |
| 120 |
255635_at |
AT4G00720
|
ASKTHETA, SHAGGY-LIKE PROTEIN KINASE THETA, ATSK32, shaggy-like protein kinase 32, SK32, shaggy-like protein kinase 32 |
-0.257 |
-0.020 |
| 121 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.247 |
-0.125 |
| 122 |
267062_at |
AT2G32600
|
[hydroxyproline-rich glycoprotein family protein] |
-0.246 |
0.099 |
| 123 |
251404_at |
AT3G60310
|
unknown |
-0.246 |
-0.062 |
| 124 |
251861_at |
AT3G54810
|
BME3, BLUE MICROPYLAR END 3, BME3-ZF, BLUE MICROPYLAR END 3-ZINC FINGER, GATA8, GATA TRANSCRIPTION FACTOR 8 |
-0.246 |
-0.052 |
| 125 |
248423_at |
AT5G51670
|
unknown |
-0.242 |
-0.002 |
| 126 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.242 |
-0.046 |
| 127 |
254041_at |
AT4G25830
|
[Uncharacterised protein family (UPF0497)] |
-0.239 |
-0.046 |
| 128 |
262915_at |
AT1G59940
|
ARR3, response regulator 3 |
-0.237 |
-0.064 |
| 129 |
246144_at |
AT5G20110
|
[Dynein light chain type 1 family protein] |
-0.234 |
-0.025 |
| 130 |
257018_at |
AT3G19630
|
[Radical SAM superfamily protein] |
-0.233 |
-0.049 |
| 131 |
265011_at |
AT1G24490
|
ALB4, ALBINA 4, ARTEMIS, ARABIDOPSIS THALIANA ENVELOPE MEMBRANE INTEGRASE |
-0.229 |
-0.025 |
| 132 |
266240_at |
AT2G29580
|
MAC5B, MOS4-associated complex subunit 5B |
-0.228 |
-0.042 |
| 133 |
255309_at |
AT4G04920
|
AtSFR6, IEN1, INSENSITIVE TO EXOGENOUS NAD+ 1, MED16, MEDIATOR 16, SFR6, SENSITIVE TO FREEZING 6, YID1, YELLOW AND SENSITIVE TO IRON-DEFICIENCY 1 |
-0.227 |
-0.031 |