|
probeID |
AGICode |
Annotation |
Log2 signal ratio
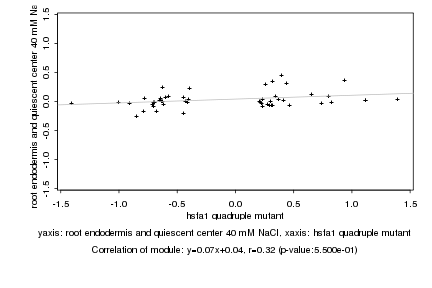
hsfa1 quadruple mutant |
Log2 signal ratio
root endodermis and quiescent center 40 mM NaCl |
| 1 |
253894_at |
AT4G27150
|
AT2S2, SESA2, seed storage albumin 2 |
1.391 |
0.048 |
| 2 |
253904_at |
AT4G27140
|
AT2S1, SESA1, seed storage albumin 1 |
1.113 |
0.018 |
| 3 |
253902_at |
AT4G27170
|
AT2S4, SESA4, seed storage albumin 4 |
1.111 |
0.030 |
| 4 |
263881_at |
AT2G21820
|
unknown |
0.931 |
0.377 |
| 5 |
248125_at |
AT5G54740
|
SESA5, seed storage albumin 5 |
0.821 |
-0.010 |
| 6 |
245233_at |
AT4G25580
|
[CAP160 protein] |
0.799 |
0.099 |
| 7 |
246552_at |
AT5G15420
|
unknown |
0.735 |
-0.023 |
| 8 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.645 |
0.136 |
| 9 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.463 |
-0.056 |
| 10 |
261247_at |
AT1G20070
|
unknown |
0.437 |
0.315 |
| 11 |
266165_at |
AT2G28190
|
CSD2, copper/zinc superoxide dismutase 2, CZSOD2, COPPER/ZINC SUPEROXIDE DISMUTASE 2 |
0.411 |
0.019 |
| 12 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
0.390 |
0.454 |
| 13 |
247640_at |
AT5G60610
|
[F-box/RNI-like superfamily protein] |
0.364 |
0.040 |
| 14 |
265856_at |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
0.341 |
0.090 |
| 15 |
263276_at |
AT2G14100
|
CYP705A13, cytochrome P450, family 705, subfamily A, polypeptide 13 |
0.318 |
-0.054 |
| 16 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.310 |
0.357 |
| 17 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.303 |
-0.068 |
| 18 |
265131_at |
AT1G23760
|
JP630, PG3, POLYGALACTURONASE 3 |
0.294 |
0.017 |
| 19 |
248178_at |
AT5G54370
|
[Late embryogenesis abundant (LEA) protein-related] |
0.288 |
-0.053 |
| 20 |
256970_at |
AT3G21090
|
ABCG15, ATP-binding cassette G15 |
0.269 |
-0.035 |
| 21 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
0.252 |
0.297 |
| 22 |
249656_at |
AT5G37130
|
[Protein prenylyltransferase superfamily protein] |
0.226 |
-0.069 |
| 23 |
258191_at |
AT3G29100
|
ATVTI13, VTI13, vesicle transport V-snare 13 |
0.224 |
0.052 |
| 24 |
250135_at |
AT5G15360
|
unknown |
0.221 |
-0.020 |
| 25 |
246417_at |
AT5G16990
|
[Zinc-binding dehydrogenase family protein] |
0.212 |
-0.012 |
| 26 |
253914_at |
AT4G27400
|
[Late embryogenesis abundant (LEA) protein-related] |
0.203 |
0.003 |
| 27 |
252462_at |
AT3G47250
|
unknown |
-1.414 |
-0.034 |
| 28 |
263023_at |
AT1G23960
|
unknown |
-1.011 |
-0.013 |
| 29 |
254585_at |
AT4G19500
|
[nucleoside-triphosphatases] |
-0.912 |
-0.021 |
| 30 |
255484_at |
AT4G02540
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.852 |
-0.250 |
| 31 |
246888_at |
AT5G26270
|
unknown |
-0.793 |
-0.157 |
| 32 |
252703_at |
AT3G43740
|
[Leucine-rich repeat (LRR) family protein] |
-0.787 |
0.059 |
| 33 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.718 |
-0.036 |
| 34 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
-0.708 |
-0.008 |
| 35 |
266516_at |
AT2G47880
|
[Glutaredoxin family protein] |
-0.706 |
-0.076 |
| 36 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
-0.703 |
-0.003 |
| 37 |
263673_at |
AT2G04800
|
unknown |
-0.681 |
-0.158 |
| 38 |
245451_at |
AT4G16890
|
BAL, BALL, SNC1, SUPPRESSOR OF NPR1-1, CONSTITUTIVE 1 |
-0.660 |
0.025 |
| 39 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.647 |
0.059 |
| 40 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-0.636 |
0.012 |
| 41 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
-0.630 |
0.014 |
| 42 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.628 |
0.251 |
| 43 |
262458_at |
AT1G11280
|
[S-locus lectin protein kinase family protein] |
-0.620 |
-0.036 |
| 44 |
260701_at |
AT1G32330
|
ATHSFA1D, heat shock transcription factor A1D, HSFA1D, heat shock transcription factor A1D |
-0.601 |
0.079 |
| 45 |
251142_at |
AT5G01015
|
unknown |
-0.576 |
0.089 |
| 46 |
246818_at |
AT5G27270
|
EMB976, EMBRYO DEFECTIVE 976 |
-0.452 |
0.081 |
| 47 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.451 |
-0.202 |
| 48 |
253514_at |
AT4G31805
|
POLAR, POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION |
-0.430 |
0.002 |
| 49 |
254663_at |
AT4G18290
|
KAT2, potassium channel in Arabidopsis thaliana 2 |
-0.414 |
-0.004 |
| 50 |
260664_at |
AT1G19510
|
ATRL5, RAD-like 5, RL5, RAD-like 5, RSM4, RADIALIS-LIKE SANT/MYB 4 |
-0.404 |
0.035 |
| 51 |
254836_at |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
-0.395 |
0.232 |