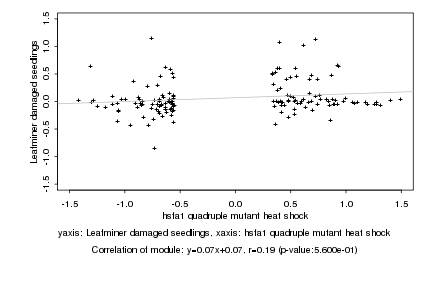
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
hsfa1 quadruple mutant heat shock |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
266393_at |
AT2G41260
|
ATM17, M17 |
1.489 |
0.037 |
| 2 |
253894_at |
AT4G27150
|
AT2S2, SESA2, seed storage albumin 2 |
1.397 |
0.021 |
| 3 |
253767_at |
AT4G28520
|
CRC, CRUCIFERIN C, CRU3, cruciferin 3 |
1.305 |
-0.059 |
| 4 |
256354_at |
AT1G54870
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.267 |
-0.016 |
| 5 |
254095_at |
AT4G25140
|
OLE1, OLEOSIN 1, OLEO1, oleosin 1 |
1.267 |
-0.052 |
| 6 |
255048_at |
AT4G09600
|
GASA3, GAST1 protein homolog 3 |
1.255 |
-0.052 |
| 7 |
250468_at |
AT5G10120
|
[Ethylene insensitive 3 family protein] |
1.187 |
-0.051 |
| 8 |
265891_at |
AT2G15010
|
[Plant thionin] |
1.169 |
-0.009 |
| 9 |
247061_at |
AT5G66780
|
unknown |
1.096 |
-0.007 |
| 10 |
246552_at |
AT5G15420
|
unknown |
1.075 |
-0.020 |
| 11 |
257130_at |
AT3G20210
|
DELTA-VPE, delta vacuolar processing enzyme, DELTAVPE, DELTA VACUOLAR PROCESSING ENZYME |
1.054 |
-0.006 |
| 12 |
263175_at |
AT1G05510
|
OBAP1a, Oil Body-Associated Protein 1a |
0.989 |
0.067 |
| 13 |
263753_at |
AT2G21490
|
LEA, dehydrin LEA |
0.974 |
0.002 |
| 14 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.927 |
0.654 |
| 15 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.919 |
0.667 |
| 16 |
258327_at |
AT3G22640
|
PAP85 |
0.916 |
-0.038 |
| 17 |
245233_at |
AT4G25580
|
[CAP160 protein] |
0.896 |
0.032 |
| 18 |
251731_at |
AT3G56350
|
[Iron/manganese superoxide dismutase family protein] |
0.889 |
-0.047 |
| 19 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.883 |
-0.037 |
| 20 |
259167_at |
AT3G01570
|
[Oleosin family protein] |
0.875 |
0.052 |
| 21 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.862 |
0.487 |
| 22 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
0.858 |
-0.335 |
| 23 |
260994_at |
AT1G12130
|
[Flavin-binding monooxygenase family protein] |
0.846 |
-0.056 |
| 24 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.841 |
0.016 |
| 25 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
0.815 |
0.047 |
| 26 |
253930_at |
AT4G26740
|
AtCLO1, ATPXG1, ARABIDOPSIS THALIANA PEROXYGENASE 1, ATS1, seed gene 1, CLO1, CALEOSIN1 |
0.767 |
0.040 |
| 27 |
265327_at |
AT2G18210
|
unknown |
0.759 |
0.117 |
| 28 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
0.738 |
-0.053 |
| 29 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.736 |
0.401 |
| 30 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.720 |
1.140 |
| 31 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.717 |
0.094 |
| 32 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
0.690 |
-0.159 |
| 33 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.682 |
0.474 |
| 34 |
266900_at |
AT2G34610
|
unknown |
0.681 |
0.009 |
| 35 |
264635_at |
AT1G65500
|
unknown |
0.666 |
0.405 |
| 36 |
251652_at |
AT3G57380
|
[Glycosyltransferase family 61 protein] |
0.663 |
0.162 |
| 37 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
0.660 |
-0.013 |
| 38 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
0.628 |
-0.103 |
| 39 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.612 |
1.018 |
| 40 |
261305_at |
AT1G48470
|
GLN1;5, glutamine synthetase 1;5 |
0.609 |
0.044 |
| 41 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.592 |
0.001 |
| 42 |
264494_at |
AT1G27461
|
unknown |
0.585 |
-0.020 |
| 43 |
259961_at |
AT1G53700
|
PK3AT, PROTEIN KINASE 3 ARABIDOPSIS THALIANA, WAG1, WAG 1 |
0.559 |
-0.018 |
| 44 |
256114_at |
AT1G16850
|
unknown |
0.550 |
0.455 |
| 45 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.543 |
0.610 |
| 46 |
257853_at |
AT3G12960
|
SMP1, seed maturation protein 1 |
0.535 |
0.035 |
| 47 |
251971_at |
AT3G53160
|
UGT73C7, UDP-glucosyl transferase 73C7 |
0.531 |
0.005 |
| 48 |
253532_at |
AT4G31570
|
unknown |
0.528 |
-0.138 |
| 49 |
245341_at |
AT4G16447
|
unknown |
0.526 |
-0.223 |
| 50 |
253111_at |
AT4G35940
|
unknown |
0.519 |
0.075 |
| 51 |
249072_at |
AT5G44060
|
unknown |
0.497 |
0.108 |
| 52 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.492 |
0.452 |
| 53 |
246200_at |
AT4G37240
|
unknown |
0.479 |
-0.277 |
| 54 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
0.478 |
0.003 |
| 55 |
263138_at |
AT1G65090
|
unknown |
0.478 |
0.019 |
| 56 |
256416_at |
AT3G11050
|
ATFER2, ferritin 2, FER2, ferritin 2 |
0.470 |
0.120 |
| 57 |
263881_at |
AT2G21820
|
unknown |
0.455 |
0.408 |
| 58 |
247640_at |
AT5G60610
|
[F-box/RNI-like superfamily protein] |
0.440 |
-0.068 |
| 59 |
265856_at |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
0.423 |
-0.014 |
| 60 |
267478_at |
AT2G02700
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.415 |
-0.005 |
| 61 |
265039_at |
AT1G04000
|
unknown |
0.415 |
-0.188 |
| 62 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.411 |
-0.057 |
| 63 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.404 |
0.246 |
| 64 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.399 |
0.013 |
| 65 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.395 |
1.073 |
| 66 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.394 |
0.609 |
| 67 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.389 |
-0.016 |
| 68 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.380 |
0.606 |
| 69 |
248981_at |
AT5G45110
|
ATNPR3, NPR3, NPR1-like protein 3 |
0.374 |
0.218 |
| 70 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
0.366 |
0.014 |
| 71 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
0.356 |
-0.416 |
| 72 |
258610_at |
AT3G02875
|
ILR1, IAA-LEUCINE RESISTANT 1 |
0.355 |
0.527 |
| 73 |
245697_at |
AT5G04200
|
AtMC9, metacaspase 9, AtMCP2f, metacaspase 2f, MC9, metacaspase 9, MCP2f, metacaspase 2f |
0.345 |
-0.073 |
| 74 |
263892_at |
AT2G36890
|
ATMYB38, MYB DOMAIN PROTEIN 38, BIT1, BLUE INSENSITIVE TRAIT 1, MYB38, MYB DOMAIN PROTEIN 38, RAX2, REGULATOR OF AXILLARY MERISTEMS 2 |
0.343 |
0.015 |
| 75 |
266737_at |
AT2G47140
|
AtSDR5, SDR5, short-chain dehydrogenase reductase 5 |
0.342 |
0.315 |
| 76 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.334 |
0.512 |
| 77 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.329 |
0.496 |
| 78 |
258695_at |
AT3G09640
|
APX1B, ASCORBATE PEROXIDASE 1B, APX2, ascorbate peroxidase 2 |
-1.428 |
0.034 |
| 79 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
-1.316 |
0.642 |
| 80 |
252462_at |
AT3G47250
|
unknown |
-1.305 |
-0.005 |
| 81 |
248215_at |
AT5G53680
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-1.289 |
0.035 |
| 82 |
251130_at |
AT5G01180
|
AtNPF8.2, ATPTR5, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 5, NPF8.2, NRT1/ PTR family 8.2, PTR5, peptide transporter 5 |
-1.256 |
-0.089 |
| 83 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-1.184 |
-0.105 |
| 84 |
258827_at |
AT3G07150
|
unknown |
-1.115 |
-0.051 |
| 85 |
245243_at |
AT1G44414
|
unknown |
-1.114 |
0.108 |
| 86 |
254674_at |
AT4G18450
|
[Integrase-type DNA-binding superfamily protein] |
-1.075 |
-0.028 |
| 87 |
256518_at |
AT1G66080
|
unknown |
-1.068 |
-0.351 |
| 88 |
250036_at |
AT5G18340
|
[ARM repeat superfamily protein] |
-1.064 |
-0.163 |
| 89 |
254263_at |
AT4G23493
|
unknown |
-1.060 |
-0.172 |
| 90 |
257322_at |
ATMG01180
|
ORF111B |
-1.032 |
0.051 |
| 91 |
263023_at |
AT1G23960
|
unknown |
-1.002 |
0.040 |
| 92 |
264402_at |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
-0.957 |
-0.424 |
| 93 |
255485_at |
AT4G02550
|
unknown |
-0.925 |
0.378 |
| 94 |
264899_at |
AT1G23130
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.911 |
-0.022 |
| 95 |
255651_at |
AT4G00940
|
AtDOF4.1, DOF4.1, DNA binding with one finger 4.1, ITD1, INTERCELLULAR TRAFFICKING DOF 1 |
-0.894 |
-0.097 |
| 96 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
-0.880 |
0.082 |
| 97 |
254062_at |
AT4G25380
|
AtSAP10, Arabidopsis thaliana stress-associated protein 10, SAP10, stress-associated protein 10 |
-0.873 |
0.052 |
| 98 |
250013_at |
AT5G18040
|
unknown |
-0.865 |
-0.035 |
| 99 |
267190_at |
AT2G44170
|
ATNMT2, ARABIDOPSIS N-MYRISTOYLTRANSFERASE 2, NMT2, N-myristoyltransferase 2 |
-0.857 |
-0.057 |
| 100 |
253689_at |
AT4G29770
|
unknown |
-0.846 |
-0.047 |
| 101 |
245272_at |
AT4G17250
|
unknown |
-0.842 |
0.015 |
| 102 |
256331_at |
AT1G76880
|
[Duplicated homeodomain-like superfamily protein] |
-0.832 |
-0.274 |
| 103 |
246888_at |
AT5G26270
|
unknown |
-0.799 |
0.273 |
| 104 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.788 |
-0.423 |
| 105 |
251522_at |
AT3G59430
|
unknown |
-0.766 |
-0.118 |
| 106 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
-0.766 |
1.153 |
| 107 |
250162_at |
AT5G15250
|
ATFTSH6, FTSH6, FTSH protease 6 |
-0.752 |
-0.045 |
| 108 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
-0.745 |
-0.323 |
| 109 |
251724_at |
AT3G56250
|
unknown |
-0.735 |
0.033 |
| 110 |
254574_at |
AT4G19430
|
unknown |
-0.734 |
-0.848 |
| 111 |
262458_at |
AT1G11280
|
[S-locus lectin protein kinase family protein] |
-0.718 |
-0.129 |
| 112 |
260701_at |
AT1G32330
|
ATHSFA1D, heat shock transcription factor A1D, HSFA1D, heat shock transcription factor A1D |
-0.712 |
-0.039 |
| 113 |
249139_at |
AT5G43170
|
AZF3, zinc-finger protein 3, ZF3, zinc-finger protein 3 |
-0.709 |
0.291 |
| 114 |
259540_at |
AT1G20640
|
NLP4, NIN-like protein 4 |
-0.699 |
-0.176 |
| 115 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
-0.694 |
0.043 |
| 116 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
-0.691 |
-0.081 |
| 117 |
261081_at |
AT1G07350
|
SR45a, serine/arginine rich-like protein 45a |
-0.688 |
-0.210 |
| 118 |
246612_at |
AT5G35320
|
unknown |
-0.687 |
0.002 |
| 119 |
265660_at |
AT2G25470
|
AtRLP21, receptor like protein 21, RLP21, receptor like protein 21 |
-0.681 |
0.467 |
| 120 |
248774_at |
AT5G47830
|
unknown |
-0.671 |
-0.070 |
| 121 |
256563_at |
AT3G29780
|
RALFL27, ralf-like 27 |
-0.669 |
-0.042 |
| 122 |
262014_at |
AT1G35660
|
unknown |
-0.663 |
-0.257 |
| 123 |
258603_at |
AT3G02990
|
ATHSFA1E, heat shock transcription factor A1E, HSFA1E, heat shock transcription factor A1E |
-0.662 |
0.122 |
| 124 |
266516_at |
AT2G47880
|
[Glutaredoxin family protein] |
-0.654 |
0.088 |
| 125 |
262656_at |
AT1G14200
|
[RING/U-box superfamily protein] |
-0.640 |
-0.105 |
| 126 |
256363_at |
AT1G66510
|
[AAR2 protein family] |
-0.640 |
-0.136 |
| 127 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
-0.640 |
0.635 |
| 128 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
-0.636 |
-0.143 |
| 129 |
259268_at |
AT3G01070
|
AtENODL16, ENODL16, early nodulin-like protein 16 |
-0.635 |
-0.020 |
| 130 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
-0.625 |
-0.188 |
| 131 |
259529_at |
AT1G12400
|
[Nucleotide excision repair, TFIIH, subunit TTDA] |
-0.607 |
0.005 |
| 132 |
253987_at |
AT4G26270
|
PFK3, phosphofructokinase 3 |
-0.600 |
0.151 |
| 133 |
245313_at |
AT4G15420
|
[Ubiquitin fusion degradation UFD1 family protein] |
-0.599 |
0.043 |
| 134 |
247510_at |
AT5G62030
|
[diphthamide synthesis DPH2 family protein] |
-0.597 |
-0.003 |
| 135 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
-0.593 |
0.582 |
| 136 |
254211_at |
AT4G23570
|
SGT1A |
-0.589 |
-0.131 |
| 137 |
258794_at |
AT3G04710
|
TPR10, tetratricopeptide repeat 10 |
-0.586 |
-0.113 |
| 138 |
257822_at |
AT3G25230
|
ATFKBP62, FKBP62, FK506 BINDING PROTEIN 62, ROF1, rotamase FKBP 1 |
-0.584 |
-0.244 |
| 139 |
263673_at |
AT2G04800
|
unknown |
-0.582 |
0.015 |
| 140 |
250899_at |
AT5G03340
|
AtCDC48C, cell division cycle 48C |
-0.578 |
-0.038 |
| 141 |
264465_at |
AT1G10230
|
ASK18, SKP1-like 18, SK18, SKP1-like 18 |
-0.578 |
0.005 |
| 142 |
263873_at |
AT2G21860
|
[violaxanthin de-epoxidase-related] |
-0.577 |
-0.181 |
| 143 |
246733_at |
AT5G27660
|
DEG14, degradation of periplasmic proteins 14 |
-0.575 |
-0.020 |
| 144 |
262450_at |
AT1G11320
|
unknown |
-0.573 |
-0.039 |
| 145 |
246018_at |
AT5G10695
|
unknown |
-0.571 |
0.515 |
| 146 |
266011_at |
AT2G37440
|
[DNAse I-like superfamily protein] |
-0.567 |
0.067 |
| 147 |
261839_at |
AT1G16040
|
unknown |
-0.565 |
0.117 |
| 148 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
-0.564 |
-0.161 |
| 149 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
-0.563 |
-0.380 |
| 150 |
262641_at |
AT1G62730
|
[Terpenoid synthases superfamily protein] |
-0.563 |
0.097 |
| 151 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.562 |
0.446 |
| 152 |
245903_at |
AT5G11100
|
ATSYTD, NTMC2T2.2, NTMC2TYPE2.2, SYT4, synaptotagmin 4, SYTD |
-0.561 |
-0.089 |
| 153 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
-0.553 |
-0.070 |