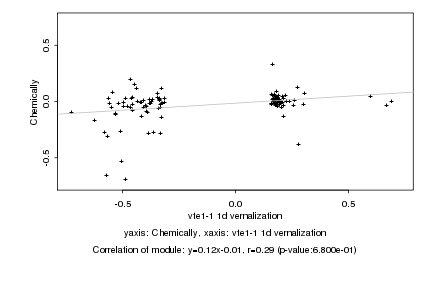
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
vte1-1 1d vernalization |
Log2 signal ratio
Chemically |
| 1 |
249456_at |
AT5G39410
|
[Saccharopine dehydrogenase ] |
0.690 |
0.004 |
| 2 |
254440_at |
AT4G21020
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.668 |
-0.028 |
| 3 |
264888_at |
AT1G23070
|
unknown |
0.596 |
0.045 |
| 4 |
247953_at |
AT5G57260
|
CYP71B10, cytochrome P450, family 71, subfamily B, polypeptide 10 |
0.304 |
0.080 |
| 5 |
257451_at |
AT1G05690
|
BT3, BTB and TAZ domain protein 3 |
0.298 |
-0.025 |
| 6 |
247170_at |
AT5G65530
|
[Protein kinase superfamily protein] |
0.277 |
-0.374 |
| 7 |
262120_at |
AT1G02950
|
ATGSTF4, glutathione S-transferase F4, GST31, GLUTATHIONE S-TRANSFERASE 31, GSTF4, glutathione S-transferase F4 |
0.274 |
0.128 |
| 8 |
251012_at |
AT5G02580
|
unknown |
0.258 |
0.015 |
| 9 |
251490_at |
AT3G59490
|
unknown |
0.256 |
-0.027 |
| 10 |
258036_at |
AT3G21210
|
[zinc ion binding] |
0.239 |
0.006 |
| 11 |
256898_at |
AT3G24650
|
ABI3, ABA INSENSITIVE 3, ABI3, ABSCISIC ACID INSENSITIVE 3, SIS10, SUGAR INSENSITIVE 10 |
0.223 |
0.008 |
| 12 |
260994_at |
AT1G12130
|
[Flavin-binding monooxygenase family protein] |
0.223 |
0.006 |
| 13 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
0.220 |
0.059 |
| 14 |
245555_at |
AT4G15390
|
[HXXXD-type acyl-transferase family protein] |
0.211 |
-0.133 |
| 15 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
0.210 |
-0.029 |
| 16 |
252309_at |
AT3G49340
|
[Cysteine proteinases superfamily protein] |
0.209 |
0.029 |
| 17 |
247805_at |
AT5G58180
|
ATYKT62, YKT62 |
0.206 |
0.052 |
| 18 |
254147_at |
AT4G24270
|
EMB140, EMBRYO DEFECTIVE 140 |
0.205 |
-0.006 |
| 19 |
263138_at |
AT1G65090
|
unknown |
0.203 |
-0.000 |
| 20 |
262719_at |
AT1G43590
|
unknown |
0.202 |
-0.045 |
| 21 |
256412_at |
AT3G11220
|
ELO1, ELONGATA 1 |
0.199 |
-0.013 |
| 22 |
264720_at |
AT1G70080
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.195 |
-0.023 |
| 23 |
246896_at |
AT5G25550
|
[Leucine-rich repeat (LRR) family protein] |
0.194 |
0.023 |
| 24 |
249716_at |
AT5G35770
|
SAP, STERILE APETALA |
0.191 |
0.008 |
| 25 |
256632_at |
AT3G28330
|
[F-box family protein-related] |
0.190 |
0.062 |
| 26 |
259022_at |
AT3G07420
|
ATNS2, ASPARAGINYL-TRNA SYNTHETASE 2, NS2, asparaginyl-tRNA synthetase 2, SYNC2, SYNTHETASE C2, SYNC2_ARATH, ASPARAGINYL-TRNA SYNTHETASE 2 |
0.189 |
0.015 |
| 27 |
254854_at |
AT4G12130
|
[Glycine cleavage T-protein family] |
0.189 |
0.011 |
| 28 |
244909_at |
ATMG00740
|
ORF100A |
0.187 |
0.007 |
| 29 |
254672_at |
AT4G18420
|
unknown |
0.186 |
-0.042 |
| 30 |
257012_at |
AT3G26120
|
terminal EAR1-like 1 |
0.186 |
-0.043 |
| 31 |
252731_at |
AT3G43130
|
[pseudogene] |
0.184 |
0.012 |
| 32 |
264345_at |
AT1G11915
|
unknown |
0.182 |
0.037 |
| 33 |
257401_at |
AT1G23550
|
SRO2, similar to RCD one 2 |
0.182 |
-0.007 |
| 34 |
250590_at |
AT5G07710
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.182 |
0.032 |
| 35 |
253299_at |
AT4G33800
|
unknown |
0.181 |
-0.035 |
| 36 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
0.180 |
0.042 |
| 37 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
0.179 |
0.091 |
| 38 |
260210_at |
AT1G74420
|
ATFUT3, FUT3, fucosyltransferase 3 |
0.178 |
0.044 |
| 39 |
252614_at |
AT3G45220
|
[Serine protease inhibitor (SERPIN) family protein] |
0.177 |
0.018 |
| 40 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
0.175 |
-0.010 |
| 41 |
262265_at |
AT1G42460
|
[cysteine-type peptidases] |
0.175 |
-0.023 |
| 42 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.174 |
-0.029 |
| 43 |
263103_at |
AT2G05290
|
unknown |
0.174 |
0.047 |
| 44 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.173 |
0.034 |
| 45 |
257806_at |
AT3G18670
|
[Ankyrin repeat family protein] |
0.172 |
0.009 |
| 46 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.170 |
0.069 |
| 47 |
248148_at |
AT5G54930
|
[AT hook motif-containing protein] |
0.169 |
0.022 |
| 48 |
258874_at |
AT3G03230
|
[alpha/beta-Hydrolases superfamily protein] |
0.169 |
-0.006 |
| 49 |
261075_at |
AT1G07280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.169 |
-0.009 |
| 50 |
255090_at |
AT4G09360
|
[NB-ARC domain-containing disease resistance protein] |
0.167 |
0.033 |
| 51 |
252351_at |
AT3G48210
|
unknown |
0.166 |
0.031 |
| 52 |
254349_at |
AT4G22250
|
[RING/U-box superfamily protein] |
0.166 |
0.005 |
| 53 |
250214_at |
AT5G13870
|
EXGT-A4, endoxyloglucan transferase A4, XTH5, xyloglucan endotransglucosylase/hydrolase 5 |
0.165 |
0.020 |
| 54 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.162 |
0.335 |
| 55 |
253253_at |
AT4G34750
|
SAUR49, SMALL AUXIN UPREGULATED RNA 49 |
0.162 |
0.019 |
| 56 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.160 |
0.061 |
| 57 |
257584_at |
AT1G54955
|
unknown |
0.158 |
-0.009 |
| 58 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.157 |
-0.011 |
| 59 |
256217_at |
AT1G56320
|
unknown |
0.156 |
-0.024 |
| 60 |
256588_at |
AT3G28790
|
unknown |
0.156 |
0.064 |
| 61 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
-0.729 |
-0.096 |
| 62 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
-0.625 |
-0.165 |
| 63 |
260344_at |
AT1G69240
|
ATMES15, ARABIDOPSIS THALIANA METHYL ESTERASE 15, MES15, methyl esterase 15, RHS9, ROOT HAIR SPECIFIC 9 |
-0.584 |
-0.269 |
| 64 |
255632_at |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
-0.571 |
-0.650 |
| 65 |
266336_at |
AT2G32270
|
ZIP3, zinc transporter 3 precursor |
-0.567 |
-0.310 |
| 66 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
-0.565 |
0.031 |
| 67 |
246004_at |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
-0.560 |
-0.011 |
| 68 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.550 |
-0.050 |
| 69 |
254327_at |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.547 |
0.089 |
| 70 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
-0.533 |
-0.098 |
| 71 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
-0.532 |
-0.107 |
| 72 |
249482_at |
AT5G38980
|
unknown |
-0.522 |
-0.016 |
| 73 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
-0.509 |
-0.264 |
| 74 |
249756_at |
AT5G24313
|
unknown |
-0.508 |
-0.529 |
| 75 |
258183_at |
AT3G21550
|
AtDMP2, Arabidopsis thaliana DUF679 domain membrane protein 2, DMP2, DUF679 domain membrane protein 2 |
-0.500 |
-0.006 |
| 76 |
245318_at |
AT4G16980
|
[arabinogalactan-protein family] |
-0.499 |
-0.042 |
| 77 |
252327_at |
AT3G48740
|
AtSWEET11, SWEET11 |
-0.488 |
0.035 |
| 78 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
-0.487 |
-0.685 |
| 79 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
-0.481 |
-0.037 |
| 80 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.468 |
0.198 |
| 81 |
258184_at |
AT3G21510
|
AHP1, histidine-containing phosphotransmitter 1 |
-0.465 |
-0.049 |
| 82 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.463 |
0.030 |
| 83 |
260867_at |
AT1G43790
|
TED6, tracheary element differentiation-related 6 |
-0.460 |
-0.026 |
| 84 |
254551_at |
AT4G19840
|
ATPP2-A1, phloem protein 2-A1, ATPP2A-1, phloem protein 2-A1, PP2-A1, phloem protein 2-A1 |
-0.458 |
0.044 |
| 85 |
250958_at |
AT5G03260
|
LAC11, laccase 11 |
-0.456 |
-0.077 |
| 86 |
259329_at |
AT3G16360
|
AHP4, HPT phosphotransmitter 4 |
-0.451 |
0.154 |
| 87 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
-0.440 |
0.123 |
| 88 |
252199_at |
AT3G50270
|
[HXXXD-type acyl-transferase family protein] |
-0.438 |
0.001 |
| 89 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
-0.422 |
-0.005 |
| 90 |
263475_at |
AT2G31945
|
unknown |
-0.420 |
-0.004 |
| 91 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
-0.418 |
-0.133 |
| 92 |
264007_at |
AT2G21140
|
ATPRP2, proline-rich protein 2, PRP2, proline-rich protein 2 |
-0.410 |
-0.053 |
| 93 |
247648_at |
AT5G60020
|
ATLAC17, LAC17, laccase 17 |
-0.408 |
0.013 |
| 94 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.401 |
-0.035 |
| 95 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.398 |
-0.040 |
| 96 |
266849_at |
AT2G25940
|
ALPHA-VPE, alpha-vacuolar processing enzyme, ALPHAVPE |
-0.395 |
-0.083 |
| 97 |
255587_at |
AT4G01480
|
AtPPa5, pyrophosphorylase 5, PPa5, pyrophosphorylase 5 |
-0.393 |
-0.091 |
| 98 |
259195_at |
AT3G01730
|
unknown |
-0.388 |
-0.280 |
| 99 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.384 |
-0.013 |
| 100 |
263845_at |
AT2G37040
|
ATPAL1, PAL1, PHE ammonia lyase 1 |
-0.383 |
0.024 |
| 101 |
251785_at |
AT3G55130
|
ABCG19, ATP-binding cassette G19, ATWBC19, white-brown complex homolog 19, WBC19, white-brown complex homolog 19 |
-0.380 |
-0.012 |
| 102 |
245792_at |
AT1G32100
|
ATPRR1, PRR1, pinoresinol reductase 1 |
-0.376 |
0.003 |
| 103 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.368 |
0.024 |
| 104 |
251723_at |
AT3G56230
|
[BTB/POZ domain-containing protein] |
-0.367 |
-0.270 |
| 105 |
259358_at |
AT1G13250
|
GATL3, galacturonosyltransferase-like 3 |
-0.346 |
0.040 |
| 106 |
247304_at |
AT5G63850
|
AAP4, amino acid permease 4 |
-0.346 |
0.080 |
| 107 |
248028_at |
AT5G55620
|
unknown |
-0.341 |
-0.057 |
| 108 |
249887_at |
AT5G22310
|
unknown |
-0.340 |
0.024 |
| 109 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
-0.340 |
0.013 |
| 110 |
249010_at |
AT5G44580
|
unknown |
-0.337 |
0.030 |
| 111 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.335 |
-0.047 |
| 112 |
250683_x_at |
AT5G06640
|
EXT10, extensin 10 |
-0.335 |
-0.284 |
| 113 |
245765_at |
AT1G33600
|
[Leucine-rich repeat (LRR) family protein] |
-0.333 |
-0.026 |
| 114 |
267580_at |
AT2G41990
|
unknown |
-0.331 |
-0.137 |
| 115 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.329 |
0.122 |
| 116 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
-0.329 |
-0.011 |
| 117 |
251058_at |
AT5G01790
|
unknown |
-0.327 |
-0.010 |
| 118 |
250231_at |
AT5G13910
|
LEP, LEAFY PETIOLE |
-0.317 |
0.029 |
| 119 |
262288_at |
AT1G70760
|
CRR23, CHLORORESPIRATORY REDUCTION 23, NdhL, NADH dehydrogenase-like complex L |
-0.315 |
-0.005 |