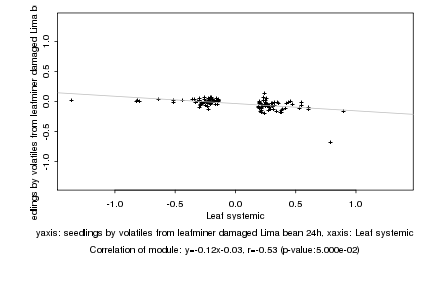
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Leaf systemic |
Log2 signal ratio
seedlings by volatiles from leafminer damaged Lima bean 24h |
| 1 |
253643_at |
AT4G29780
|
unknown |
0.889 |
-0.160 |
| 2 |
252346_at |
AT3G48650
|
[pseudogene] |
0.785 |
-0.668 |
| 3 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.605 |
-0.121 |
| 4 |
246495_at |
AT5G16200
|
[50S ribosomal protein-related] |
0.604 |
-0.090 |
| 5 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.546 |
-0.053 |
| 6 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
0.544 |
-0.011 |
| 7 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.530 |
-0.116 |
| 8 |
264000_at |
AT2G22500
|
ATPUMP5, PLANT UNCOUPLING MITOCHONDRIAL PROTEIN 5, DIC1, DICARBOXYLATE CARRIER 1, UCP5, uncoupling protein 5 |
0.468 |
-0.034 |
| 9 |
263182_at |
AT1G05575
|
unknown |
0.451 |
0.009 |
| 10 |
245755_at |
AT1G35210
|
unknown |
0.436 |
-0.013 |
| 11 |
263379_at |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
0.423 |
-0.020 |
| 12 |
267069_at |
AT2G41010
|
ATCAMBP25, calmodulin (CAM)-binding protein of 25 kDa, CAMBP25, calmodulin (CAM)-binding protein of 25 kDa |
0.413 |
-0.116 |
| 13 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.387 |
-0.117 |
| 14 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.377 |
-0.143 |
| 15 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.374 |
-0.180 |
| 16 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.372 |
-0.170 |
| 17 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.355 |
-0.018 |
| 18 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.344 |
-0.002 |
| 19 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.339 |
-0.154 |
| 20 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.324 |
-0.063 |
| 21 |
267289_at |
AT2G23770
|
LYK4, LysM-containing receptor-like kinase 4 |
0.319 |
-0.008 |
| 22 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
0.309 |
-0.121 |
| 23 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.305 |
-0.067 |
| 24 |
253323_at |
AT4G33920
|
APD5, Arabidopsis Pp2c clade D 5 |
0.302 |
-0.024 |
| 25 |
256872_at |
AT3G26490
|
[Phototropic-responsive NPH3 family protein] |
0.294 |
-0.022 |
| 26 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
0.291 |
-0.055 |
| 27 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
0.286 |
-0.122 |
| 28 |
247055_at |
AT5G66740
|
unknown |
0.282 |
-0.089 |
| 29 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
0.275 |
-0.058 |
| 30 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
0.274 |
-0.143 |
| 31 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.267 |
-0.060 |
| 32 |
246289_at |
AT3G56880
|
[VQ motif-containing protein] |
0.265 |
-0.035 |
| 33 |
250335_at |
AT5G11650
|
[alpha/beta-Hydrolases superfamily protein] |
0.258 |
-0.067 |
| 34 |
259812_at |
AT1G49840
|
unknown |
0.257 |
-0.029 |
| 35 |
265452_at |
AT2G46510
|
AIB, ABA-inducible BHLH-type transcription factor, ATAIB, ABA-inducible BHLH-type transcription factor, JAM1, JA-associated MYC2-like 1 |
0.256 |
0.057 |
| 36 |
256755_at |
AT3G25600
|
[Calcium-binding EF-hand family protein] |
0.251 |
-0.049 |
| 37 |
259792_at |
AT1G29690
|
CAD1, constitutively activated cell death 1 |
0.250 |
-0.062 |
| 38 |
264389_at |
AT1G11960
|
[ERD (early-responsive to dehydration stress) family protein] |
0.246 |
-0.000 |
| 39 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
0.246 |
-0.039 |
| 40 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
0.242 |
-0.072 |
| 41 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.242 |
0.022 |
| 42 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
0.242 |
-0.060 |
| 43 |
258122_at |
AT3G14570
|
ATGSL04, glucan synthase-like 4, atgsl4, gsl04, GSL04, glucan synthase-like 4 |
0.241 |
-0.039 |
| 44 |
261983_at |
AT1G33770
|
[Protein kinase superfamily protein] |
0.240 |
0.145 |
| 45 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
0.240 |
-0.198 |
| 46 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.232 |
0.071 |
| 47 |
252391_at |
AT3G47860
|
CHL, chloroplastic lipocalin |
0.225 |
0.024 |
| 48 |
266798_at |
AT2G22850
|
AtbZIP6, basic leucine-zipper 6, bZIP6, basic leucine-zipper 6 |
0.217 |
-0.069 |
| 49 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.215 |
-0.181 |
| 50 |
247464_at |
AT5G62070
|
IQD23, IQ-domain 23 |
0.211 |
-0.034 |
| 51 |
263918_at |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
0.211 |
-0.113 |
| 52 |
257057_at |
AT3G15310
|
unknown |
0.210 |
-0.134 |
| 53 |
253165_at |
AT4G35320
|
unknown |
0.209 |
-0.181 |
| 54 |
263797_at |
AT2G24570
|
ATWRKY17, WRKY17, WRKY DNA-binding protein 17 |
0.209 |
-0.099 |
| 55 |
263845_at |
AT2G37040
|
ATPAL1, PAL1, PHE ammonia lyase 1 |
0.207 |
-0.025 |
| 56 |
251281_at |
AT3G61640
|
AGP20, arabinogalactan protein 20, AtAGP20, Arabinogalactan protein 20 |
0.205 |
-0.021 |
| 57 |
267260_at |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
0.201 |
-0.159 |
| 58 |
266757_at |
AT2G46940
|
unknown |
0.197 |
-0.105 |
| 59 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
0.197 |
-0.006 |
| 60 |
266464_at |
AT2G47800
|
ABCC4, ATP-binding cassette C4, ATMRP4, multidrug resistance-associated protein 4, EST3, MRP4, multidrug resistance-associated protein 4 |
0.194 |
0.015 |
| 61 |
251896_at |
AT3G54390
|
[sequence-specific DNA binding transcription factors] |
0.193 |
-0.024 |
| 62 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.191 |
-0.099 |
| 63 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
0.187 |
-0.071 |
| 64 |
267459_at |
AT2G33850
|
unknown |
-1.366 |
0.023 |
| 65 |
259009_at |
AT3G09260
|
BGLU23, LEB, LONG ER BODY, PSR3.1, PYK10 |
-0.827 |
0.003 |
| 66 |
249813_at |
AT5G23940
|
DCR, DEFECTIVE IN CUTICULAR RIDGES, EMB3009, EMBRYO DEFECTIVE 3009, PEL3, PERMEABLE LEAVES3 |
-0.814 |
0.019 |
| 67 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.799 |
0.010 |
| 68 |
253753_at |
AT4G29030
|
[Putative membrane lipoprotein] |
-0.642 |
0.042 |
| 69 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
-0.520 |
-0.013 |
| 70 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
-0.518 |
0.026 |
| 71 |
258059_at |
AT3G29035
|
ANAC059, Arabidopsis NAC domain containing protein 59, ATNAC3, NAC domain containing protein 3, NAC3, NAC domain containing protein 3, ORS1, ORE1 SISTER1 |
-0.443 |
0.025 |
| 72 |
262819_at |
AT1G11600
|
CYP77B1, cytochrome P450, family 77, subfamily B, polypeptide 1 |
-0.358 |
0.038 |
| 73 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
-0.340 |
0.039 |
| 74 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
-0.336 |
-0.011 |
| 75 |
266797_at |
AT2G22840
|
AtGRF1, growth-regulating factor 1, GRF1, growth-regulating factor 1 |
-0.317 |
0.012 |
| 76 |
254964_at |
AT4G11080
|
3xHMG-box1, 3xHigh Mobility Group-box1 |
-0.302 |
0.053 |
| 77 |
256062_at |
AT1G07090
|
LSH6, LIGHT SENSITIVE HYPOCOTYLS 6 |
-0.301 |
0.027 |
| 78 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.300 |
-0.093 |
| 79 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.298 |
-0.037 |
| 80 |
250937_at |
AT5G03230
|
unknown |
-0.290 |
-0.062 |
| 81 |
263914_at |
AT2G36400
|
AtGRF3, growth-regulating factor 3, GRF3, growth-regulating factor 3 |
-0.285 |
-0.042 |
| 82 |
262096_at |
AT1G56010
|
anac021, Arabidopsis NAC domain containing protein 21, ANAC022, Arabidopsis NAC domain containing protein 22, NAC1, NAC domain containing protein 1 |
-0.284 |
-0.011 |
| 83 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.283 |
-0.017 |
| 84 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
-0.274 |
-0.025 |
| 85 |
259686_at |
AT1G63100
|
[GRAS family transcription factor] |
-0.261 |
0.034 |
| 86 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.261 |
-0.003 |
| 87 |
267096_at |
AT2G38180
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.260 |
0.067 |
| 88 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
-0.253 |
-0.052 |
| 89 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.252 |
0.014 |
| 90 |
258385_at |
AT3G15510
|
ANAC056, Arabidopsis NAC domain containing protein 56, ATNAC2, NAC domain containing protein 2, NAC2, NAC domain containing protein 2, NARS1, NAC-REGULATED SEED MORPHOLOGY 1 |
-0.243 |
-0.016 |
| 91 |
262170_at |
AT1G74940
|
unknown |
-0.238 |
-0.005 |
| 92 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
-0.238 |
0.003 |
| 93 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
-0.235 |
-0.078 |
| 94 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.234 |
-0.082 |
| 95 |
266223_at |
AT2G28790
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.231 |
0.008 |
| 96 |
248100_at |
AT5G55180
|
[O-Glycosyl hydrolases family 17 protein] |
-0.228 |
-0.013 |
| 97 |
252464_at |
AT3G47160
|
[RING/U-box superfamily protein] |
-0.228 |
0.023 |
| 98 |
265128_at |
AT1G30860
|
[RING/U-box superfamily protein] |
-0.227 |
0.066 |
| 99 |
254954_at |
AT4G10910
|
unknown |
-0.227 |
-0.118 |
| 100 |
259912_at |
AT1G72670
|
iqd8, IQ-domain 8 |
-0.226 |
-0.014 |
| 101 |
252036_at |
AT3G52070
|
unknown |
-0.222 |
0.005 |
| 102 |
266912_at |
AT2G45900
|
TRM13, TON1 Recruiting Motif 13 |
-0.222 |
0.019 |
| 103 |
255980_at |
AT1G33970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.218 |
-0.002 |
| 104 |
246505_at |
AT5G16250
|
unknown |
-0.216 |
-0.017 |
| 105 |
247812_at |
AT5G58390
|
[Peroxidase superfamily protein] |
-0.214 |
0.067 |
| 106 |
245702_at |
AT5G04220
|
ATSYTC, NTMC2T1.3, NTMC2TYPE1.3, SYT3, synaptotagmin 3, SYTC |
-0.213 |
0.056 |
| 107 |
251358_at |
AT3G61160
|
[Protein kinase superfamily protein] |
-0.210 |
-0.033 |
| 108 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
-0.208 |
0.014 |
| 109 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.203 |
-0.014 |
| 110 |
253065_at |
AT4G37740
|
AtGRF2, growth-regulating factor 2, GRF2, growth-regulating factor 2 |
-0.203 |
0.076 |
| 111 |
257381_at |
AT2G37950
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.197 |
0.037 |
| 112 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.194 |
-0.004 |
| 113 |
262236_at |
AT1G48330
|
unknown |
-0.192 |
0.037 |
| 114 |
261364_at |
AT1G53140
|
DRP5A, Dynamin related protein 5A |
-0.191 |
-0.014 |
| 115 |
265720_at |
AT2G40110
|
[Yippee family putative zinc-binding protein] |
-0.184 |
0.023 |
| 116 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
-0.183 |
0.004 |
| 117 |
246239_at |
AT4G36790
|
[Major facilitator superfamily protein] |
-0.183 |
0.007 |
| 118 |
250565_at |
AT5G08000
|
E13L3, glucan endo-1,3-beta-glucosidase-like protein 3, PDCB2, PLASMODESMATA CALLOSE-BINDING PROTEIN 2 |
-0.178 |
0.009 |
| 119 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
-0.173 |
-0.048 |
| 120 |
258034_at |
AT3G21300
|
[RNA methyltransferase family protein] |
-0.160 |
0.033 |
| 121 |
259598_at |
AT1G27980
|
ATDPL1, DPL1, dihydrosphingosine phosphate lyase |
-0.158 |
0.055 |
| 122 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
-0.150 |
-0.035 |
| 123 |
247599_at |
AT5G60880
|
BASL, BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE |
-0.148 |
0.027 |
| 124 |
259080_at |
AT3G04910
|
ATWNK1, WNK1, with no lysine (K) kinase 1, ZIK4 |
-0.147 |
0.003 |
| 125 |
251778_at |
AT3G55660
|
ATROPGEF6, ROPGEF6, ROP (rho of plants) guanine nucleotide exchange factor 6 |
-0.146 |
0.042 |
| 126 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
-0.146 |
0.019 |