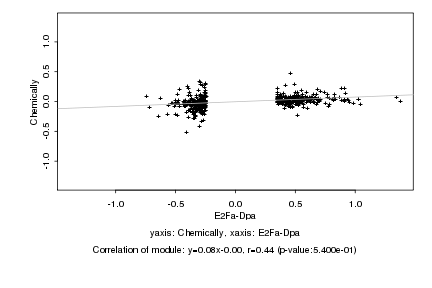
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
E2Fa-Dpa |
Log2 signal ratio
Chemically |
| 1 |
256120_at |
AT1G18130
|
[Class II aaRS and biotin synthetases superfamily protein] |
1.375 |
0.010 |
| 2 |
263955_at |
AT2G36010
|
ATE2FA, E2F3, E2F transcription factor 3 |
1.344 |
0.079 |
| 3 |
251535_at |
AT3G58540
|
unknown |
1.037 |
-0.035 |
| 4 |
251052_at |
AT5G02470
|
DPA |
1.024 |
0.049 |
| 5 |
260459_at |
AT1G68240
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
1.021 |
0.048 |
| 6 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.982 |
-0.033 |
| 7 |
254443_at |
AT4G21070
|
ATBRCA1, ARABIDOPSIS THALIANA BREAST CANCER SUSCEPTIBILITY1, BRCA1, breast cancer susceptibility1 |
0.952 |
-0.005 |
| 8 |
254728_at |
AT4G13690
|
unknown |
0.943 |
0.032 |
| 9 |
251772_at |
AT3G55920
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.930 |
0.041 |
| 10 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
0.917 |
0.145 |
| 11 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
0.910 |
0.223 |
| 12 |
253869_at |
AT4G27510
|
unknown |
0.907 |
0.021 |
| 13 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.906 |
0.017 |
| 14 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.904 |
0.043 |
| 15 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.881 |
0.227 |
| 16 |
251035_at |
AT5G02220
|
SMR4, SIAMESE-RELATED 4 |
0.879 |
0.025 |
| 17 |
258490_at |
AT3G02670
|
[Glycine-rich protein family] |
0.868 |
0.072 |
| 18 |
263535_at |
AT2G24970
|
unknown |
0.828 |
0.036 |
| 19 |
249836_at |
AT5G23420
|
HMGB6, high-mobility group box 6 |
0.820 |
0.078 |
| 20 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.819 |
0.132 |
| 21 |
258242_at |
AT3G27640
|
[Transducin/WD40 repeat-like superfamily protein] |
0.811 |
0.025 |
| 22 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.805 |
0.036 |
| 23 |
250588_at |
AT5G07660
|
SMC6A, structural maintenance of chromosomes 6A |
0.784 |
-0.036 |
| 24 |
246315_at |
AT3G56870
|
unknown |
0.780 |
0.051 |
| 25 |
266616_at |
AT2G29680
|
ATCDC6, CDC6, cell division control 6 |
0.771 |
-0.069 |
| 26 |
250823_at |
AT5G05180
|
unknown |
0.762 |
0.121 |
| 27 |
248481_at |
AT5G50930
|
[Histone superfamily protein] |
0.762 |
0.075 |
| 28 |
262036_at |
AT1G35530
|
At-FANCM, FANCM, Fanconi anemia complementation group M |
0.747 |
-0.032 |
| 29 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.738 |
0.160 |
| 30 |
253733_at |
AT4G29170
|
ATMND1 |
0.709 |
0.045 |
| 31 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.706 |
0.171 |
| 32 |
253313_at |
AT4G33870
|
[Peroxidase superfamily protein] |
0.700 |
0.029 |
| 33 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.694 |
0.007 |
| 34 |
267173_at |
AT2G37560
|
ATORC2, ORIGIN RECOGNITION COMPLEX SECOND LARGEST SUBUNIT 2, ORC2, origin recognition complex second largest subunit 2 |
0.690 |
0.063 |
| 35 |
261027_at |
AT1G01340
|
ACBK1, ATCNGC10, cyclic nucleotide gated channel 10, CNGC10, cyclic nucleotide gated channel 10 |
0.683 |
0.034 |
| 36 |
262201_at |
AT2G01120
|
ATORC4, ORIGIN RECOGNITION COMPLEX SUBUNIT 4, ORC4, origin recognition complex subunit 4 |
0.683 |
-0.007 |
| 37 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.682 |
0.205 |
| 38 |
253817_at |
AT4G28310
|
unknown |
0.674 |
0.042 |
| 39 |
250078_at |
AT5G16690
|
ATORC3, ORC3, origin recognition complex subunit 3 |
0.673 |
-0.034 |
| 40 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.672 |
0.128 |
| 41 |
251378_at |
AT3G60660
|
unknown |
0.661 |
0.083 |
| 42 |
256426_at |
AT1G33420
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.657 |
0.052 |
| 43 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
0.655 |
0.053 |
| 44 |
249379_at |
AT5G40460
|
unknown |
0.652 |
-0.002 |
| 45 |
257524_at |
AT3G01330
|
DEL3, DP-E2F-like protein 3, E2FF, E2L2, E2F-LIKE 2 |
0.650 |
-0.012 |
| 46 |
246898_at |
AT5G25580
|
unknown |
0.647 |
0.118 |
| 47 |
245869_at |
AT1G26330
|
[DNA binding] |
0.644 |
0.012 |
| 48 |
245762_at |
AT1G27880
|
[DEAD/DEAH box RNA helicase family protein ] |
0.643 |
0.076 |
| 49 |
263041_at |
AT1G23320
|
TAR1, tryptophan aminotransferase related 1 |
0.629 |
0.034 |
| 50 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.627 |
0.076 |
| 51 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
0.625 |
0.060 |
| 52 |
258702_at |
AT3G09730
|
unknown |
0.621 |
0.052 |
| 53 |
246132_at |
AT5G20850
|
ATRAD51, RAD51 |
0.621 |
0.033 |
| 54 |
257167_at |
AT3G24340
|
CHR40, chromatin remodeling 40 |
0.620 |
-0.010 |
| 55 |
258898_at |
AT3G05740
|
RECQI1, RECQ helicase l1 |
0.617 |
0.003 |
| 56 |
247056_at |
AT5G66750
|
ATDDM1, CHA1, CHR01, CHROMATIN REMODELING 1, CHR1, chromatin remodeling 1, DDM1, DECREASED DNA METHYLATION 1, SOM1, SOMNIFEROUS 1, SOM4 |
0.613 |
0.073 |
| 57 |
254625_at |
AT4G18470
|
SNI1, SUPPRESSOR OF NPR1-1, INDUCIBLE 1 |
0.612 |
0.061 |
| 58 |
254565_at |
AT4G19130
|
[Replication factor-A protein 1-related] |
0.610 |
0.019 |
| 59 |
257201_at |
AT3G23740
|
unknown |
0.601 |
-0.115 |
| 60 |
258630_at |
AT3G02820
|
[zinc knuckle (CCHC-type) family protein] |
0.597 |
0.033 |
| 61 |
264604_at |
AT1G04650
|
unknown |
0.596 |
0.074 |
| 62 |
256798_at |
AT3G18630
|
ATUNG, uracil dna glycosylase, UNG, uracil dna glycosylase |
0.594 |
0.023 |
| 63 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
0.593 |
-0.008 |
| 64 |
262106_at |
AT1G02970
|
ATWEE1, WEE1, WEE1 kinase homolog |
0.593 |
0.052 |
| 65 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
0.588 |
0.062 |
| 66 |
264635_at |
AT1G65500
|
unknown |
0.585 |
0.165 |
| 67 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.577 |
0.098 |
| 68 |
249788_at |
AT5G24330
|
ATXR6, ARABIDOPSIS TRITHORAX-RELATED PROTEIN 6, SDG34, SET DOMAIN PROTEIN 34 |
0.575 |
0.035 |
| 69 |
245380_at |
AT4G17760
|
[damaged DNA binding] |
0.568 |
0.023 |
| 70 |
247528_at |
AT5G61460
|
ATRAD18, MIM, hypersensitive to MMS, irradiation and MMC, SMC6B, STRUCTURAL MAINTENANCE OF CHROMOSOMES 6B |
0.565 |
0.057 |
| 71 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.564 |
0.054 |
| 72 |
256507_at |
AT1G75150
|
unknown |
0.562 |
-0.046 |
| 73 |
252539_at |
AT3G45730
|
unknown |
0.560 |
-0.096 |
| 74 |
247028_at |
AT5G67100
|
ICU2, INCURVATA2 |
0.559 |
0.007 |
| 75 |
265311_at |
AT2G20250
|
unknown |
0.558 |
0.006 |
| 76 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.557 |
0.126 |
| 77 |
248668_at |
AT5G48720
|
XRI, X-RAY INDUCED TRANSCRIPT, XRI1, X-RAY INDUCED TRANSCRIPT 1 |
0.555 |
0.097 |
| 78 |
260945_at |
AT1G05950
|
unknown |
0.555 |
0.053 |
| 79 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.553 |
-0.018 |
| 80 |
265348_at |
AT2G22600
|
[RNA-binding KH domain-containing protein] |
0.552 |
0.084 |
| 81 |
261741_at |
AT1G47870
|
ATE2F2, ATE2FC, ARABIDOPSIS THALIANA HOMOLOG OF E2F C, E2FC |
0.551 |
0.070 |
| 82 |
258138_at |
AT3G24495
|
ATMSH7, ARABIDOPSIS THALIANA MUTS HOMOLOG 7, MSH6-2, MUTS HOMOLOG 6-2, MSH7, MUTS homolog 7 |
0.550 |
0.031 |
| 83 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.550 |
0.187 |
| 84 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.546 |
0.067 |
| 85 |
250616_at |
AT5G07280
|
EMS1, EXCESS MICROSPOROCYTES1, EXS, EXTRA SPOROGENOUS CELLS |
0.542 |
-0.004 |
| 86 |
254287_at |
AT4G22960
|
unknown |
0.539 |
0.043 |
| 87 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.537 |
-0.016 |
| 88 |
251014_at |
AT5G02520
|
KNL2, KINETOCHORE NULL 2 |
0.532 |
0.006 |
| 89 |
265097_at |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
0.531 |
0.044 |
| 90 |
247301_at |
AT5G63920
|
AtTOP3alpha, TOP3A, topoisomerase 3alpha |
0.530 |
0.002 |
| 91 |
263681_at |
AT1G26840
|
ATORC6, ARABIDOPSIS THALIANA ORIGIN RECOGNITION COMPLEX PROTEIN 6, ORC6, origin recognition complex protein 6 |
0.529 |
0.024 |
| 92 |
247317_at |
AT5G64060
|
anac103, NAC domain containing protein 103, NAC103, NAC domain containing protein 103 |
0.529 |
0.023 |
| 93 |
251147_at |
AT3G63480
|
[ATP binding microtubule motor family protein] |
0.528 |
0.048 |
| 94 |
254946_at |
AT4G10950
|
[SGNH hydrolase-type esterase superfamily protein] |
0.527 |
-0.023 |
| 95 |
255562_at |
AT4G02070
|
ATMSH6, ARABIDOPSIS THALIANA MUTS HOMOLOG 6, MSH6, MUTS homolog 6, MSH6-1, MUTS HOMOLOG 6-1 |
0.526 |
0.060 |
| 96 |
253175_at |
AT4G35050
|
MSI3, MULTICOPY SUPPRESSOR OF IRA1 3, NFC3, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 3 |
0.526 |
-0.039 |
| 97 |
264969_at |
AT1G67320
|
EMB2813, EMBRYO DEFECTIVE 2813 |
0.525 |
0.093 |
| 98 |
266234_at |
AT2G02350
|
AtPP2-B9, Phloem protein 2-B9, SKIP3, SKP1 interacting partner 3 |
0.524 |
0.023 |
| 99 |
264600_at |
AT1G04730
|
CTF18, CHROMOSOME TRANSMISSION FIDELITY 18 |
0.524 |
-0.001 |
| 100 |
263515_at |
AT2G21640
|
unknown |
0.523 |
-0.057 |
| 101 |
261000_at |
AT1G26540
|
[Agenet domain-containing protein] |
0.521 |
0.046 |
| 102 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.520 |
0.110 |
| 103 |
266959_at |
AT2G07690
|
MCM5, MINICHROMOSOME MAINTENANCE 5 |
0.520 |
0.029 |
| 104 |
253599_at |
AT4G30860
|
ASHR3, ASH1-related 3, SDG4, SET domain group 4 |
0.520 |
0.094 |
| 105 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.520 |
0.113 |
| 106 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.519 |
0.057 |
| 107 |
248940_at |
AT5G45400
|
ATRPA70C, RPA70C |
0.517 |
0.090 |
| 108 |
256320_at |
AT3G12170
|
[Chaperone DnaJ-domain superfamily protein] |
0.517 |
0.001 |
| 109 |
247559_at |
AT5G61070
|
ATHDA18, A. THALIANA HISTONE DEACETYLASE OF THE RPD3/HDA1 SUPERFAMILY 18, HDA18, histone deacetylase of the RPD3/HDA1 superfamily 18 |
0.516 |
-0.016 |
| 110 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.516 |
0.163 |
| 111 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.515 |
-0.229 |
| 112 |
256082_at |
AT1G20720
|
[RAD3-like DNA-binding helicase protein] |
0.511 |
0.029 |
| 113 |
266511_at |
AT2G47680
|
[zinc finger (CCCH type) helicase family protein] |
0.509 |
-0.043 |
| 114 |
245585_at |
AT4G14970
|
unknown |
0.507 |
-0.016 |
| 115 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.505 |
0.025 |
| 116 |
261330_at |
AT1G44900
|
ATMCM2, MCM2, MINICHROMOSOME MAINTENANCE 2 |
0.505 |
0.093 |
| 117 |
246790_at |
AT5G27610
|
ALY1, ALWAYS EARLY 1, ATALY1, ARABIDOPSIS THALIANA ALWAYS EARLY 1 |
0.504 |
0.026 |
| 118 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
0.503 |
0.117 |
| 119 |
250914_at |
AT5G03780
|
TRFL10, TRF-like 10 |
0.500 |
0.015 |
| 120 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.500 |
-0.042 |
| 121 |
265437_at |
AT2G20980
|
MCM10, minichromosome maintenance 10 |
0.499 |
0.042 |
| 122 |
256284_at |
AT3G12530
|
PSF2 |
0.498 |
0.001 |
| 123 |
258011_at |
AT3G19510
|
HAT3.1 |
0.498 |
0.074 |
| 124 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.497 |
0.165 |
| 125 |
260524_at |
AT2G47230
|
unknown |
0.496 |
0.032 |
| 126 |
246141_at |
AT5G19920
|
[Transducin/WD40 repeat-like superfamily protein] |
0.492 |
0.049 |
| 127 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.487 |
0.300 |
| 128 |
260192_at |
AT1G67630
|
EMB2814, EMBRYO DEFECTIVE 2814, POLA2, DNA polymerase alpha 2 |
0.487 |
0.043 |
| 129 |
256429_at |
AT3G11040
|
AtENGase85B, ENGase85B, Endo-beta-N-acetyglucosaminidase 85B |
0.487 |
0.012 |
| 130 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.485 |
0.117 |
| 131 |
248938_at |
AT5G45780
|
[Leucine-rich repeat protein kinase family protein] |
0.485 |
0.010 |
| 132 |
263130_at |
AT1G78650
|
POLD3 |
0.485 |
0.101 |
| 133 |
251543_at |
AT3G58770
|
unknown |
0.484 |
0.069 |
| 134 |
256409_at |
AT1G66620
|
[Protein with RING/U-box and TRAF-like domains] |
0.483 |
0.033 |
| 135 |
259224_at |
AT3G07800
|
AtTK1a, TK1a, thymidine kinase 1a |
0.482 |
-0.014 |
| 136 |
252361_at |
AT3G48490
|
unknown |
0.482 |
-0.007 |
| 137 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.478 |
0.019 |
| 138 |
261401_at |
AT1G79640
|
[Protein kinase superfamily protein] |
0.478 |
0.051 |
| 139 |
250369_at |
AT5G11300
|
CYC2BAT, CYC3B, mitotic-like cyclin 3B from Arabidopsis, CYCA2;2, CYCLIN A2;2 |
0.478 |
0.083 |
| 140 |
253240_at |
AT4G34510
|
KCS17, 3-ketoacyl-CoA synthase 17 |
0.476 |
-0.076 |
| 141 |
255481_at |
AT4G02460
|
AtPMS1, PMS1, POSTMEIOTIC SEGREGATION 1 |
0.474 |
-0.091 |
| 142 |
255874_at |
AT2G40550
|
ETG1, E2F target gene 1 |
0.472 |
0.035 |
| 143 |
248632_at |
AT5G49010
|
EMB2812, EMBRYO DEFECTIVE 2812, SLD5, SYNTHETIC LETHALITY WITH DPB11-1 5 |
0.470 |
-0.010 |
| 144 |
264293_at |
AT1G78770
|
APC6, anaphase promoting complex 6 |
0.469 |
0.081 |
| 145 |
257579_at |
AT3G11000
|
[DCD (Development and Cell Death) domain protein] |
0.465 |
-0.031 |
| 146 |
258459_at |
AT3G17290
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 3.1e-27 P-value blast match to GB:CAA29005 ORFa of Maize Ac (hAT-element) (Zea mays)] |
0.463 |
0.061 |
| 147 |
263612_at |
AT2G16440
|
MCM4, MINICHROMOSOME MAINTENANCE 4 |
0.458 |
0.082 |
| 148 |
254173_at |
AT4G24580
|
REN1, ROP1 ENHANCER 1 |
0.457 |
-0.095 |
| 149 |
248641_at |
AT5G49110
|
unknown |
0.456 |
-0.024 |
| 150 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.455 |
0.477 |
| 151 |
245458_at |
AT4G16970
|
[Protein kinase superfamily protein] |
0.453 |
0.000 |
| 152 |
264091_at |
AT1G79110
|
BRG2, BOI-related gene 2 |
0.452 |
0.096 |
| 153 |
257725_at |
AT3G18524
|
ATMSH2, MSH2, MUTS homolog 2 |
0.452 |
0.003 |
| 154 |
251863_at |
AT3G54870
|
ARK1, ARMADILLO REPEAT-CONTAINING KINESIN 1, AtKINUc, Arabidopsis thaliana KINESIN Ungrouped clade, gene A, CAE1, CA-ROP2 ENHANCER 1, MRH2, MORPHOGENESIS OF ROOT HAIR 2 |
0.450 |
-0.062 |
| 155 |
258697_at |
AT3G09660
|
AtMCM8, MCM8, minichromosome maintenance 8 |
0.448 |
0.059 |
| 156 |
251986_at |
AT3G53310
|
[AP2/B3-like transcriptional factor family protein] |
0.448 |
0.009 |
| 157 |
256268_at |
AT3G12280
|
ATRBR1, RETINOBLASTOMA-RELATED PROTEIN 1, RB, RB1, RETINOBLASTOMA 1, RBR, RETINOBLASTOMA-RELATED, RBR1, retinoblastoma-related 1 |
0.447 |
0.005 |
| 158 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
0.444 |
0.011 |
| 159 |
247887_at |
AT5G57880
|
ATPRD2, ARABIDOPSIS THALIANA PUTATIVE RECOMBINATION INITIATION DEFECTS 2, MPS1, MULTIPOLAR SPINDLE 1, PRD2, PUTATIVE RECOMBINATION INITIATION DEFECTS 2 |
0.442 |
-0.074 |
| 160 |
249028_at |
AT5G44740
|
POLH, Y-family DNA polymerase H |
0.442 |
0.012 |
| 161 |
253110_at |
AT4G35930
|
AtFBS4, FBS4, F-box stress induced 4 |
0.441 |
0.024 |
| 162 |
266412_at |
AT2G38720
|
MAP65-5, microtubule-associated protein 65-5 |
0.439 |
0.023 |
| 163 |
257929_at |
AT3G16980
|
NRPB9A, NRPD9A, NRPE9A |
0.437 |
0.008 |
| 164 |
248139_at |
AT5G54970
|
unknown |
0.437 |
0.070 |
| 165 |
263264_at |
AT2G38810
|
HTA8, histone H2A 8 |
0.436 |
0.015 |
| 166 |
251850_at |
AT3G54650
|
FBL17 |
0.436 |
-0.044 |
| 167 |
250672_at |
AT5G06910
|
ATJ6, J-domain protein 6 |
0.435 |
0.027 |
| 168 |
260506_at |
AT1G47210
|
CYCA3;2, cyclin-dependent protein kinase 3;2 |
0.434 |
0.004 |
| 169 |
251955_at |
AT3G53680
|
[Acyl-CoA N-acyltransferase with RING/FYVE/PHD-type zinc finger domain] |
0.429 |
-0.006 |
| 170 |
257765_at |
AT3G23020
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.427 |
0.024 |
| 171 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
0.425 |
0.077 |
| 172 |
253567_at |
AT4G31230
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.424 |
0.038 |
| 173 |
266603_at |
AT2G46040
|
[ARID/BRIGHT DNA-binding domain] |
0.422 |
0.030 |
| 174 |
260159_at |
AT1G79890
|
[RAD3-like DNA-binding helicase protein] |
0.421 |
-0.018 |
| 175 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
0.421 |
0.008 |
| 176 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.421 |
0.073 |
| 177 |
266754_at |
AT2G46980
|
ASY3, ASYNAPTIC 3, AtASY3, Arabidopsis thaliana ASYNAPTIC 3 |
0.421 |
0.058 |
| 178 |
248675_at |
AT5G48820
|
ICK6, inhibitor/interactor with cyclin-dependent kinase, KRP3, KIP-RELATED PROTEIN 3 |
0.419 |
0.023 |
| 179 |
260909_at |
AT1G02670
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.417 |
-0.051 |
| 180 |
257494_at |
AT1G32375
|
[F-box/RNI-like/FBD-like domains-containing protein] |
0.414 |
0.092 |
| 181 |
256887_at |
AT3G15150
|
ATMMS21, A. THALIANA METHYL METHANE SULFONATE SENSITIVITY 21, HPY2, HIGH PLOIDY2, MMS21, METHYL METHANE SULFONATE SENSITIVITY 2 |
0.414 |
-0.040 |
| 182 |
263882_at |
AT2G21790
|
ATRNR1, RIBONUCLEOTIDE REDUCTASE LARGE SUBUNIT 1, CLS8, CRINKLY LEAVES 8, DPD2, defective in pollen organelle DNA degradation 2, R1, RIBONUCLEOTIDE REDUCTASE 1, RNR1, ribonucleotide reductase 1 |
0.414 |
0.079 |
| 183 |
264630_at |
AT1G65470
|
FAS1, FASCIATA 1, FUGU2, FUGU 2, NFB2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP B |
0.414 |
0.050 |
| 184 |
245194_at |
AT1G67820
|
[Protein phosphatase 2C family protein] |
0.413 |
0.014 |
| 185 |
249276_at |
AT5G41880
|
POLA3, POLA4 |
0.413 |
0.042 |
| 186 |
248279_at |
AT5G52910
|
ATIM, TIMELESS |
0.412 |
0.272 |
| 187 |
252770_at |
AT3G42860
|
[zinc knuckle (CCHC-type) family protein] |
0.411 |
0.020 |
| 188 |
258235_at |
AT3G27620
|
AOX1C, alternative oxidase 1C |
0.410 |
0.034 |
| 189 |
250212_at |
AT5G13960
|
KYP, KRYPTONITE, SDG33, SET DOMAIN PROTEIN 33, SUVH4, SU(VAR)3-9 homolog 4 |
0.408 |
0.050 |
| 190 |
247200_at |
AT5G65120
|
unknown |
0.407 |
0.028 |
| 191 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.405 |
0.077 |
| 192 |
257159_at |
AT3G24400
|
AtPERK2, proline-rich extensin-like receptor kinase 2, PERK2, proline-rich extensin-like receptor kinase 2 |
0.405 |
0.018 |
| 193 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
0.404 |
-0.102 |
| 194 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
0.404 |
0.050 |
| 195 |
267190_at |
AT2G44170
|
ATNMT2, ARABIDOPSIS N-MYRISTOYLTRANSFERASE 2, NMT2, N-myristoyltransferase 2 |
0.404 |
0.037 |
| 196 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.401 |
0.145 |
| 197 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.399 |
-0.008 |
| 198 |
262410_at |
AT1G34770
|
unknown |
0.397 |
-0.014 |
| 199 |
265383_at |
AT2G16780
|
MSI02, MSI2, MULTICOPY SUPPRESSOR OF IRA1 2, NFC02, NFC2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 2 |
0.397 |
0.099 |
| 200 |
245147_at |
AT2G45280
|
ATRAD51C, RAS associated with diabetes protein 51C, RAD51C, RAS associated with diabetes protein 51C |
0.394 |
0.009 |
| 201 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.394 |
-0.012 |
| 202 |
265705_at |
AT2G03410
|
[Mo25 family protein] |
0.392 |
-0.011 |
| 203 |
265695_at |
AT2G24490
|
ATRPA2, ATRPA32A, ROR1, SUPPRESSOR OF ROS1, RPA2, replicon protein A2, RPA32A |
0.390 |
0.076 |
| 204 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.389 |
-0.038 |
| 205 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
0.388 |
-0.005 |
| 206 |
259252_at |
AT3G07610
|
IBM1, increase in bonsai methylation 1 |
0.386 |
0.045 |
| 207 |
254349_at |
AT4G22250
|
[RING/U-box superfamily protein] |
0.385 |
0.005 |
| 208 |
254350_at |
AT4G22280
|
[F-box/RNI-like superfamily protein] |
0.385 |
-0.010 |
| 209 |
257164_at |
AT3G24320
|
ATMSH1, CHM, CHLOROPLAST MUTATOR, CHM1, MSH1, MUTL protein homolog 1 |
0.385 |
0.024 |
| 210 |
252785_at |
AT3G42660
|
[transducin family protein / WD-40 repeat family protein] |
0.384 |
0.021 |
| 211 |
259780_at |
AT1G29630
|
[5'-3' exonuclease family protein] |
0.384 |
-0.025 |
| 212 |
257086_at |
AT3G20490
|
unknown protein; Has 754 Blast hits to 165 proteins in 64 species: Archae - 0; Bacteria - 48; Metazoa - 26; Fungi - 25; Plants - 36; Viruses - 0; Other Eukaryotes - 619 (source: NCBI BLink). |
0.383 |
0.029 |
| 213 |
257334_at |
ATMG01370
|
ORF111D |
0.383 |
-0.004 |
| 214 |
263446_at |
AT2G31650
|
ATX1, homologue of trithorax, SDG27, SET DOMAIN PROTEIN 27 |
0.383 |
-0.007 |
| 215 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
0.381 |
0.076 |
| 216 |
255613_at |
AT4G01270
|
[RING/U-box superfamily protein] |
0.381 |
-0.022 |
| 217 |
252351_at |
AT3G48210
|
unknown |
0.380 |
0.031 |
| 218 |
261058_at |
AT1G01370
|
CENH3, CENTROMERIC HISTONE H3, HTR12 |
0.380 |
0.091 |
| 219 |
246413_at |
AT1G77310
|
unknown |
0.380 |
0.050 |
| 220 |
257809_at |
AT3G27060
|
ATTSO2, TSO2, TSO MEANING 'UGLY' IN CHINESE 2 |
0.380 |
0.024 |
| 221 |
248597_at |
AT5G49160
|
DDM2, DECREASED DNA METHYLATION 2, DMT01, DNA METHYLTRANSFERASE 01, DMT1, DNA METHYLTRANSFERASE 1, MET1, methyltransferase 1, MET2, METHYLTRANSFERASE 2, METI, METHYLTRANSFERASE I |
0.378 |
0.006 |
| 222 |
245236_at |
AT4G25540
|
ATMSH3, MSH3, homolog of DNA mismatch repair protein MSH3 |
0.378 |
-0.013 |
| 223 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.376 |
0.123 |
| 224 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.375 |
0.026 |
| 225 |
267576_at |
AT2G30640
|
MUG2, MUSTANG 2 |
0.374 |
-0.003 |
| 226 |
247081_at |
AT5G66130
|
ATRAD17, RADIATION SENSITIVE 17, RAD17, RADIATION SENSITIVE 17 |
0.374 |
0.045 |
| 227 |
257237_at |
AT3G14890
|
[phosphoesterase] |
0.372 |
0.074 |
| 228 |
254177_at |
AT4G23860
|
[PHD finger protein-related] |
0.371 |
0.042 |
| 229 |
267552_at |
AT2G32770
|
ATPAP13, PURPLE ACID PHOSPHATASE 13, PAP13, purple acid phosphatase 13 |
0.370 |
-0.002 |
| 230 |
247972_at |
AT5G56740
|
HAC07, HAC7, HAG02, HAG2, histone acetyltransferase of the GNAT family 2 |
0.369 |
0.025 |
| 231 |
260430_at |
AT1G68200
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.369 |
0.015 |
| 232 |
252889_at |
AT4G39380
|
unknown |
0.369 |
0.060 |
| 233 |
260276_at |
AT1G80450
|
[VQ motif-containing protein] |
0.367 |
0.065 |
| 234 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
0.366 |
0.008 |
| 235 |
259690_at |
AT1G63160
|
EMB2811, EMBRYO DEFECTIVE 2811, RFC2, replication factor C 2 |
0.365 |
0.012 |
| 236 |
248841_at |
AT5G46740
|
UBP21, ubiquitin-specific protease 21 |
0.365 |
0.027 |
| 237 |
246581_at |
AT1G31760
|
[SWIB/MDM2 domain superfamily protein] |
0.364 |
0.019 |
| 238 |
257931_at |
AT3G17030
|
[Nucleic acid-binding proteins superfamily] |
0.364 |
0.039 |
| 239 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
0.363 |
0.038 |
| 240 |
244931_at |
ATMG00630
|
ORF110B |
0.362 |
-0.020 |
| 241 |
260211_at |
AT1G74440
|
unknown |
0.360 |
0.095 |
| 242 |
254055_at |
AT4G25330
|
unknown |
0.358 |
-0.039 |
| 243 |
253518_at |
AT4G31400
|
AtCTF7, CTF7, CHROMOSOME TRANSMISSION FIDELITY 7, ECO1 |
0.357 |
-0.010 |
| 244 |
260876_at |
AT1G21460
|
AtSWEET1, SWEET1 |
0.357 |
0.020 |
| 245 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
0.356 |
-0.047 |
| 246 |
251868_at |
AT3G54490
|
RPB5E, RNA polymerase II fifth largest subunit, E |
0.354 |
0.029 |
| 247 |
247250_at |
AT5G64630
|
FAS2, FASCIATA 2, MUB3.9, NFB01, NFB1 |
0.354 |
0.071 |
| 248 |
259469_at |
AT1G19100
|
AtMORC6, DMS11, DEFECTIVE IN MERISTEM SILENCING 11, MORC6, Microrchidia 6 |
0.354 |
0.037 |
| 249 |
261898_at |
AT1G80720
|
[Mitochondrial glycoprotein family protein] |
0.353 |
0.060 |
| 250 |
266130_at |
AT2G44980
|
ASG3, ALTERED SEED GERMINATION 3, CHR10, chromatin remodeling 10 |
0.351 |
0.002 |
| 251 |
256077_at |
AT1G18090
|
[5'-3' exonuclease family protein] |
0.350 |
0.055 |
| 252 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.350 |
0.089 |
| 253 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.349 |
0.034 |
| 254 |
262124_at |
AT1G59660
|
[Nucleoporin autopeptidase] |
0.349 |
0.071 |
| 255 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.349 |
0.064 |
| 256 |
262178_at |
AT1G77860
|
KOM, KOMPEITO |
0.348 |
-0.002 |
| 257 |
249907_at |
AT5G22750
|
RAD5, RAD5A |
0.347 |
0.034 |
| 258 |
253506_at |
AT4G31980
|
unknown |
0.347 |
0.007 |
| 259 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
0.346 |
0.163 |
| 260 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.345 |
0.220 |
| 261 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.344 |
0.126 |
| 262 |
267099_at |
AT2G41460
|
ARP, apurinic endonuclease-redox protein |
0.344 |
-0.001 |
| 263 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.743 |
0.100 |
| 264 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.721 |
-0.094 |
| 265 |
255160_at |
AT4G07820
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.645 |
-0.239 |
| 266 |
256603_at |
AT3G28270
|
unknown |
-0.626 |
0.060 |
| 267 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.569 |
-0.217 |
| 268 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
-0.565 |
-0.056 |
| 269 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.537 |
-0.020 |
| 270 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.527 |
-0.022 |
| 271 |
258468_at |
AT3G06070
|
unknown |
-0.512 |
-0.068 |
| 272 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.506 |
0.026 |
| 273 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.505 |
-0.003 |
| 274 |
251142_at |
AT5G01015
|
unknown |
-0.505 |
-0.204 |
| 275 |
260012_at |
AT1G67865
|
unknown |
-0.504 |
-0.035 |
| 276 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.495 |
-0.029 |
| 277 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.491 |
-0.016 |
| 278 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
-0.491 |
0.122 |
| 279 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
-0.488 |
-0.228 |
| 280 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-0.484 |
-0.031 |
| 281 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
-0.478 |
0.033 |
| 282 |
253165_at |
AT4G35320
|
unknown |
-0.473 |
-0.079 |
| 283 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.472 |
-0.046 |
| 284 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.471 |
0.206 |
| 285 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-0.470 |
-0.042 |
| 286 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.468 |
-0.078 |
| 287 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
-0.442 |
-0.037 |
| 288 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.438 |
0.007 |
| 289 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-0.432 |
0.012 |
| 290 |
261658_at |
AT1G50040
|
unknown |
-0.430 |
-0.072 |
| 291 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.429 |
-0.012 |
| 292 |
260948_at |
AT1G06100
|
[Fatty acid desaturase family protein] |
-0.429 |
-0.024 |
| 293 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
-0.427 |
-0.043 |
| 294 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.427 |
-0.016 |
| 295 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
-0.427 |
-0.046 |
| 296 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.422 |
-0.077 |
| 297 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.421 |
0.018 |
| 298 |
254938_at |
AT4G10770
|
ATOPT7, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 7, OPT7, oligopeptide transporter 7 |
-0.420 |
-0.046 |
| 299 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
-0.409 |
-0.519 |
| 300 |
255934_at |
AT1G12740
|
CYP87A2, cytochrome P450, family 87, subfamily A, polypeptide 2 |
-0.409 |
-0.180 |
| 301 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.409 |
-0.058 |
| 302 |
253985_at |
AT4G26220
|
CCoAOMT7, caffeoyl coenzyme A ester O-methyltransferase 7 |
-0.407 |
0.257 |
| 303 |
253478_at |
AT4G32350
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.407 |
-0.002 |
| 304 |
250936_at |
AT5G03120
|
unknown |
-0.403 |
-0.049 |
| 305 |
266712_at |
AT2G46750
|
AtGulLO2, GulLO2, L -gulono-1,4-lactone ( L -GulL) oxidase 2 |
-0.400 |
0.013 |
| 306 |
258008_at |
AT3G19430
|
[late embryogenesis abundant protein-related / LEA protein-related] |
-0.398 |
-0.263 |
| 307 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
-0.394 |
-0.139 |
| 308 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
-0.394 |
0.234 |
| 309 |
260230_at |
AT1G74500
|
ATBS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, bHLH135, basic helix-loop-helix protein 135, BS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, PRE3, PACLOBUTRAZOL RESISTANCE 3, TMO7, TARGET OF MONOPTEROS 7 |
-0.394 |
0.109 |
| 310 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.390 |
-0.010 |
| 311 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
-0.389 |
-0.014 |
| 312 |
258017_at |
AT3G19370
|
unknown |
-0.389 |
-0.018 |
| 313 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.384 |
0.152 |
| 314 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.381 |
-0.058 |
| 315 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.379 |
-0.154 |
| 316 |
247461_at |
AT5G62100
|
ATBAG2, BCL-2-associated athanogene 2, BAG2, BCL-2-associated athanogene 2 |
-0.378 |
-0.124 |
| 317 |
255252_at |
AT4G04990
|
unknown |
-0.378 |
0.018 |
| 318 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.376 |
0.106 |
| 319 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.373 |
-0.082 |
| 320 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.372 |
-0.102 |
| 321 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
-0.370 |
-0.117 |
| 322 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
-0.369 |
-0.070 |
| 323 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.368 |
0.056 |
| 324 |
255876_at |
AT2G40480
|
unknown |
-0.367 |
0.045 |
| 325 |
257076_at |
AT3G19680
|
unknown |
-0.365 |
-0.041 |
| 326 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
-0.364 |
-0.014 |
| 327 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.362 |
-0.039 |
| 328 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
-0.359 |
-0.162 |
| 329 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.359 |
-0.174 |
| 330 |
257288_at |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
-0.358 |
-0.034 |
| 331 |
266191_at |
AT2G39040
|
[Peroxidase superfamily protein] |
-0.357 |
-0.164 |
| 332 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
-0.355 |
0.011 |
| 333 |
266566_at |
AT2G24040
|
[Low temperature and salt responsive protein family] |
-0.355 |
0.012 |
| 334 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.355 |
-0.124 |
| 335 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.354 |
-0.203 |
| 336 |
267057_at |
AT2G32500
|
[Stress responsive alpha-beta barrel domain protein] |
-0.353 |
-0.013 |
| 337 |
266424_at |
AT2G41330
|
[Glutaredoxin family protein] |
-0.353 |
-0.047 |
| 338 |
265539_at |
AT2G15830
|
unknown |
-0.351 |
0.030 |
| 339 |
265049_at |
AT1G52060
|
[Mannose-binding lectin superfamily protein] |
-0.351 |
-0.282 |
| 340 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-0.350 |
-0.021 |
| 341 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.350 |
-0.169 |
| 342 |
251304_at |
AT3G61990
|
OMTF3, O-MTase family 3 protein |
-0.346 |
-0.007 |
| 343 |
251353_at |
AT3G61080
|
[Protein kinase superfamily protein] |
-0.346 |
-0.012 |
| 344 |
253827_at |
AT4G28085
|
unknown |
-0.345 |
-0.087 |
| 345 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.345 |
-0.054 |
| 346 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
-0.345 |
-0.281 |
| 347 |
247359_at |
AT5G63560
|
FACT, FATTY ALCOHOL:CAFFEOYL-CoA CAFFEOYL TRANSFERASE |
-0.344 |
0.023 |
| 348 |
251107_at |
AT5G01610
|
unknown |
-0.344 |
-0.102 |
| 349 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
-0.343 |
-0.009 |
| 350 |
260493_at |
AT2G41830
|
[Uncharacterized protein] |
-0.343 |
-0.009 |
| 351 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
-0.340 |
-0.107 |
| 352 |
247470_at |
AT5G62220
|
ATGT18, glycosyltransferase 18, GT18, glycosyltransferase 18, XLT2, Xyloglucan L-side chain galactosylTransferase position 2 |
-0.339 |
-0.002 |
| 353 |
262819_at |
AT1G11600
|
CYP77B1, cytochrome P450, family 77, subfamily B, polypeptide 1 |
-0.338 |
0.003 |
| 354 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.338 |
-0.130 |
| 355 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.338 |
0.012 |
| 356 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-0.338 |
-0.062 |
| 357 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.337 |
-0.099 |
| 358 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
-0.336 |
-0.243 |
| 359 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.336 |
-0.184 |
| 360 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
-0.334 |
-0.096 |
| 361 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
-0.334 |
-0.234 |
| 362 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
-0.333 |
-0.160 |
| 363 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.331 |
-0.066 |
| 364 |
255028_at |
AT4G09890
|
unknown |
-0.328 |
0.069 |
| 365 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.328 |
0.117 |
| 366 |
248581_at |
AT5G49900
|
[Beta-glucosidase, GBA2 type family protein] |
-0.326 |
-0.036 |
| 367 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.325 |
-0.036 |
| 368 |
252383_at |
AT3G47780
|
ABCA7, ATP-binding cassette A7, ATATH6, A. THALIANA ABC2 HOMOLOG 6, ATH6, ABC2 homolog 6 |
-0.323 |
-0.017 |
| 369 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
-0.323 |
-0.052 |
| 370 |
251820_at |
AT3G55040
|
GSTL2, glutathione transferase lambda 2 |
-0.322 |
0.000 |
| 371 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
-0.322 |
-0.044 |
| 372 |
267013_at |
AT2G39180
|
ATCRR2, CCR2, CRINKLY4 related 2 |
-0.322 |
-0.039 |
| 373 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
-0.320 |
0.014 |
| 374 |
260156_at |
AT1G52880
|
ANAC018, Arabidopsis NAC domain containing protein 18, ATNAM, NAM, NO APICAL MERISTEM, NARS2, NAC-REGULATED SEED MORPHOLOGY 2 |
-0.320 |
0.018 |
| 375 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-0.320 |
-0.061 |
| 376 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.319 |
-0.125 |
| 377 |
257697_at |
AT3G12700
|
NANA, NANA |
-0.319 |
0.032 |
| 378 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.318 |
0.032 |
| 379 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
-0.317 |
-0.020 |
| 380 |
256349_at |
AT1G54890
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.317 |
-0.000 |
| 381 |
248432_at |
AT5G51390
|
unknown |
-0.316 |
-0.110 |
| 382 |
259478_at |
AT1G18980
|
[RmlC-like cupins superfamily protein] |
-0.315 |
-0.111 |
| 383 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.315 |
0.201 |
| 384 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.314 |
-0.012 |
| 385 |
252010_at |
AT3G52740
|
unknown |
-0.313 |
-0.082 |
| 386 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
-0.309 |
-0.111 |
| 387 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.309 |
-0.046 |
| 388 |
248348_at |
AT5G52190
|
[Sugar isomerase (SIS) family protein] |
-0.309 |
-0.016 |
| 389 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
-0.306 |
-0.123 |
| 390 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
-0.306 |
0.001 |
| 391 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.306 |
-0.412 |
| 392 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
-0.305 |
-0.054 |
| 393 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.305 |
0.050 |
| 394 |
265118_at |
AT1G62660
|
[Glycosyl hydrolases family 32 protein] |
-0.304 |
-0.150 |
| 395 |
251658_at |
AT3G57020
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.304 |
-0.043 |
| 396 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.301 |
0.341 |
| 397 |
249057_at |
AT5G44480
|
DUR, DEFECTIVE UGE IN ROOT |
-0.301 |
-0.095 |
| 398 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
-0.300 |
-0.066 |
| 399 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
-0.300 |
-0.020 |
| 400 |
257017_at |
AT3G19620
|
[Glycosyl hydrolase family protein] |
-0.300 |
0.046 |
| 401 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.299 |
0.330 |
| 402 |
263673_at |
AT2G04800
|
unknown |
-0.299 |
-0.172 |
| 403 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.299 |
0.016 |
| 404 |
258470_at |
AT3G06035
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.299 |
-0.087 |
| 405 |
250200_at |
AT5G14130
|
[Peroxidase superfamily protein] |
-0.298 |
0.112 |
| 406 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
-0.297 |
0.129 |
| 407 |
262666_at |
AT1G14080
|
ATFUT6, FUT6, fucosyltransferase 6 |
-0.297 |
-0.036 |
| 408 |
253193_at |
AT4G35380
|
[SEC7-like guanine nucleotide exchange family protein] |
-0.297 |
-0.088 |
| 409 |
255637_at |
AT4G00750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.296 |
0.094 |
| 410 |
266715_at |
AT2G46780
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.296 |
-0.058 |
| 411 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
-0.295 |
-0.134 |
| 412 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.295 |
-0.011 |
| 413 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.295 |
-0.048 |
| 414 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.294 |
0.060 |
| 415 |
262813_at |
AT1G11670
|
[MATE efflux family protein] |
-0.294 |
-0.083 |
| 416 |
255695_at |
AT4G00080
|
UNE11, unfertilized embryo sac 11 |
-0.293 |
-0.000 |
| 417 |
259274_at |
AT3G01220
|
ATHB20, homeobox protein 20, HB20, homeobox protein 20 |
-0.293 |
-0.076 |
| 418 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
-0.293 |
-0.179 |
| 419 |
257244_at |
AT3G24240
|
[Leucine-rich repeat receptor-like protein kinase family protein] |
-0.292 |
0.299 |
| 420 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
-0.291 |
0.041 |
| 421 |
247794_at |
AT5G58670
|
ATPLC, ARABIDOPSIS THALIANA PHOSPHOLIPASE C, ATPLC1, phospholipase C1, PLC1, phospholipase C 1, PLC1, phospholipase C1 |
-0.291 |
-0.053 |
| 422 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
-0.291 |
0.003 |
| 423 |
265734_at |
AT2G01260
|
unknown |
-0.290 |
-0.022 |
| 424 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
-0.289 |
-0.330 |
| 425 |
266278_at |
AT2G29300
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.289 |
-0.058 |
| 426 |
262105_at |
AT1G02810
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.289 |
0.005 |
| 427 |
253654_at |
AT4G30060
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.287 |
-0.020 |
| 428 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
-0.287 |
-0.094 |
| 429 |
247094_at |
AT5G66280
|
GMD1, GDP-D-mannose 4,6-dehydratase 1 |
-0.286 |
-0.190 |
| 430 |
261597_at |
AT1G49780
|
PUB26, plant U-box 26 |
-0.285 |
0.040 |
| 431 |
246885_at |
AT5G26230
|
MAKR1, membrane-associated kinase regulator 1 |
-0.284 |
-0.050 |
| 432 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.284 |
-0.001 |
| 433 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.284 |
0.094 |
| 434 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-0.284 |
-0.137 |
| 435 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.283 |
-0.164 |
| 436 |
257761_at |
AT3G23090
|
WDL3 |
-0.283 |
0.097 |
| 437 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
-0.283 |
-0.047 |
| 438 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.283 |
-0.011 |
| 439 |
250723_at |
AT5G06300
|
LOG7, LONELY GUY 7 |
-0.283 |
-0.027 |
| 440 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.282 |
0.284 |
| 441 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.282 |
-0.190 |
| 442 |
265341_at |
AT2G18360
|
[alpha/beta-Hydrolases superfamily protein] |
-0.281 |
0.016 |
| 443 |
253498_at |
AT4G31890
|
[ARM repeat superfamily protein] |
-0.281 |
-0.036 |
| 444 |
267459_at |
AT2G33850
|
unknown |
-0.280 |
-0.210 |
| 445 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
-0.280 |
0.075 |
| 446 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
-0.280 |
-0.067 |
| 447 |
254313_at |
AT4G22460
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.280 |
0.056 |
| 448 |
246036_at |
AT5G08370
|
AGAL2, alpha-galactosidase 2, AtAGAL2, alpha-galactosidase 2 |
-0.279 |
-0.027 |
| 449 |
246373_at |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
-0.279 |
-0.139 |
| 450 |
258805_at |
AT3G04010
|
[O-Glycosyl hydrolases family 17 protein] |
-0.278 |
0.017 |
| 451 |
252949_at |
AT4G38670
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.278 |
0.028 |
| 452 |
254333_at |
AT4G22753
|
ATSMO1-3, SMO1-3, sterol 4-alpha methyl oxidase 1-3 |
-0.278 |
-0.101 |
| 453 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.278 |
-0.057 |
| 454 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
-0.277 |
-0.033 |
| 455 |
262388_at |
AT1G49320
|
unknown |
-0.277 |
-0.059 |
| 456 |
258802_at |
AT3G04650
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.276 |
0.051 |
| 457 |
249242_at |
AT5G42250
|
[Zinc-binding alcohol dehydrogenase family protein] |
-0.276 |
0.011 |
| 458 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.276 |
-0.038 |
| 459 |
256097_at |
AT1G13670
|
unknown |
-0.275 |
0.002 |
| 460 |
251020_at |
AT5G02270
|
ABCI20, ATP-binding cassette I20, ATNAP9, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 9, NAP9, non-intrinsic ABC protein 9 |
-0.274 |
-0.034 |
| 461 |
255266_at |
AT4G05200
|
CRK25, cysteine-rich RLK (RECEPTOR-like protein kinase) 25 |
-0.274 |
-0.048 |
| 462 |
261224_at |
AT1G20160
|
ATSBT5.2 |
-0.272 |
-0.069 |
| 463 |
246476_at |
AT5G16730
|
[LOCATED IN: chloroplast] |
-0.272 |
0.044 |
| 464 |
248486_at |
AT5G51060
|
ATRBOHC, A. THALIANA RESPIRATORY BURST OXIDASE HOMOLOG C, RBOHC, RESPIRATORY BURST OXIDASE HOMOLOG C, RHD2, ROOT HAIR DEFECTIVE 2 |
-0.271 |
-0.197 |
| 465 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.271 |
-0.029 |
| 466 |
259429_at |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
-0.270 |
0.005 |
| 467 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.270 |
0.027 |
| 468 |
248708_at |
AT5G48560
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.269 |
0.031 |
| 469 |
253247_at |
AT4G34610
|
BLH6, BEL1-like homeodomain 6 |
-0.268 |
-0.027 |
| 470 |
252364_at |
AT3G48450
|
[RPM1-interacting protein 4 (RIN4) family protein] |
-0.268 |
-0.060 |
| 471 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
-0.268 |
-0.306 |
| 472 |
255751_at |
AT1G31950
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.267 |
-0.063 |
| 473 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
-0.267 |
0.093 |
| 474 |
265188_at |
AT1G23800
|
ALDH2B, aldehyde dehydrogenase 2B, ALDH2B7, aldehyde dehydrogenase 2B7 |
-0.266 |
-0.077 |
| 475 |
258293_at |
AT3G23430
|
ATPHO1, ARABIDOPSIS PHOSPHATE 1, PHO1, phosphate 1 |
-0.266 |
-0.065 |
| 476 |
261488_at |
AT1G14345
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.265 |
-0.031 |
| 477 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
-0.265 |
0.016 |
| 478 |
255860_at |
AT5G34940
|
AtGUS3, glucuronidase 3, GUS3, glucuronidase 3 |
-0.265 |
-0.114 |
| 479 |
267209_at |
AT2G30930
|
unknown |
-0.265 |
-0.078 |
| 480 |
263915_at |
AT2G36430
|
unknown |
-0.265 |
-0.054 |
| 481 |
246911_at |
AT5G25810
|
tny, TINY |
-0.264 |
-0.214 |
| 482 |
260432_at |
AT1G68150
|
ATWRKY9, WRKY9, WRKY DNA-binding protein 9 |
-0.264 |
-0.052 |
| 483 |
258362_at |
AT3G14280
|
unknown |
-0.264 |
-0.166 |
| 484 |
249847_at |
AT5G23210
|
SCPL34, serine carboxypeptidase-like 34 |
-0.264 |
0.010 |
| 485 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.264 |
-0.034 |
| 486 |
250952_at |
AT5G03570
|
ATIREG2, ARABIDOPSIS THALIANA IRON-REGULATED PROTEIN 2, FPN2, FERROPORTIN 2, IREG2, iron regulated 2 |
-0.264 |
-0.075 |
| 487 |
263134_at |
AT1G78570
|
ATRHM1, ARABIDOPSIS THALIANA RHAMNOSE BIOSYNTHESIS 1, RHM1, rhamnose biosynthesis 1, ROL1, REPRESSOR OF LRX1 1 |
-0.263 |
0.009 |
| 488 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.263 |
0.106 |
| 489 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
-0.263 |
0.086 |
| 490 |
252248_at |
AT3G49750
|
AtRLP44, receptor like protein 44, RLP44, receptor like protein 44 |
-0.263 |
0.012 |
| 491 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.262 |
0.167 |
| 492 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.262 |
0.021 |
| 493 |
262045_at |
AT1G80240
|
DGR1, DUF642 L-GalL responsive gene 1 |
-0.262 |
0.023 |
| 494 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
-0.262 |
0.005 |
| 495 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.261 |
-0.037 |
| 496 |
248179_at |
AT5G54380
|
THE1, THESEUS1 |
-0.261 |
-0.018 |
| 497 |
249794_at |
AT5G23530
|
AtCXE18, carboxyesterase 18, CXE18, carboxyesterase 18 |
-0.260 |
-0.065 |
| 498 |
246868_at |
AT5G26010
|
[Protein phosphatase 2C family protein] |
-0.259 |
0.003 |
| 499 |
265103_at |
AT1G31070
|
GlcNAc1pUT1, N-acetylglucosamine-1-phosphate uridylyltransferase 1 |
-0.259 |
-0.012 |
| 500 |
256600_at |
AT3G14850
|
TBL41, TRICHOME BIREFRINGENCE-LIKE 41 |
-0.259 |
0.250 |
| 501 |
255690_at |
AT4G00360
|
ATT1, ABERRANT INDUCTION OF TYPE THREE 1, CYP86A2, cytochrome P450, family 86, subfamily A, polypeptide 2 |
-0.258 |
-0.072 |
| 502 |
266196_at |
AT2G39110
|
[Protein kinase superfamily protein] |
-0.257 |
-0.137 |
| 503 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
-0.257 |
-0.149 |
| 504 |
266838_at |
AT2G25980
|
[Mannose-binding lectin superfamily protein] |
-0.256 |
-0.088 |
| 505 |
251309_at |
AT3G61220
|
SDR1, short-chain dehydrogenase/reductase 1 |
-0.256 |
-0.017 |
| 506 |
249911_at |
AT5G22740
|
ATCSLA02, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE A02, ATCSLA2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE A2, CSLA02, cellulose synthase-like A02, CSLA2, CELLULOSE SYNTHASE-LIKE A 2 |
-0.256 |
0.106 |
| 507 |
255385_at |
AT4G03610
|
[Metallo-hydrolase/oxidoreductase superfamily protein] |
-0.256 |
0.013 |
| 508 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
-0.255 |
-0.058 |
| 509 |
258367_at |
AT3G14370
|
WAG2 |
-0.255 |
0.153 |
| 510 |
266941_at |
AT2G18980
|
[Peroxidase superfamily protein] |
-0.255 |
0.317 |
| 511 |
262945_at |
AT1G79510
|
[Uncharacterized conserved protein (DUF2358)] |
-0.253 |
-0.019 |
| 512 |
252353_at |
AT3G48200
|
unknown |
-0.253 |
0.015 |
| 513 |
261032_at |
AT1G17430
|
[alpha/beta-Hydrolases superfamily protein] |
-0.253 |
0.018 |
| 514 |
264800_at |
AT1G08800
|
MyoB1, myosin binding protein 1 |
-0.253 |
-0.064 |
| 515 |
256578_at |
AT3G28200
|
[Peroxidase superfamily protein] |
-0.252 |
0.005 |
| 516 |
260007_at |
AT1G67870
|
[glycine-rich protein] |
-0.252 |
0.023 |
| 517 |
265246_at |
AT2G43050
|
ATPMEPCRD |
-0.252 |
0.192 |
| 518 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-0.252 |
-0.031 |
| 519 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.252 |
0.287 |
| 520 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
-0.252 |
0.084 |
| 521 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
-0.251 |
-0.013 |
| 522 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.251 |
0.149 |
| 523 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.249 |
0.097 |