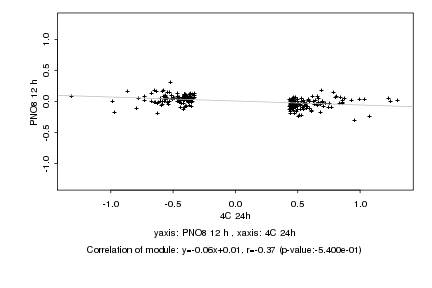
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
4C 24h |
Log2 signal ratio
PNO8 12 h |
| 1 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
1.296 |
0.032 |
| 2 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
1.239 |
0.007 |
| 3 |
256114_at |
AT1G16850
|
unknown |
1.223 |
0.064 |
| 4 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
1.072 |
-0.232 |
| 5 |
251793_at |
AT3G55580
|
[Regulator of chromosome condensation (RCC1) family protein] |
1.029 |
0.037 |
| 6 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.988 |
0.037 |
| 7 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.955 |
-0.294 |
| 8 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
0.924 |
0.032 |
| 9 |
263789_at |
AT2G24560
|
[GDSL-like Lipase/Acylhydrolase family protein] |
0.872 |
0.050 |
| 10 |
251753_at |
AT3G55760
|
unknown |
0.856 |
-0.024 |
| 11 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.854 |
0.024 |
| 12 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.853 |
-0.005 |
| 13 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.837 |
0.065 |
| 14 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.829 |
-0.031 |
| 15 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.804 |
0.093 |
| 16 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.801 |
0.069 |
| 17 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.786 |
0.148 |
| 18 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.767 |
-0.081 |
| 19 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.751 |
-0.031 |
| 20 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.741 |
-0.093 |
| 21 |
261187_at |
AT1G32860
|
[Glycosyl hydrolase superfamily protein] |
0.719 |
-0.025 |
| 22 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
0.703 |
-0.066 |
| 23 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.694 |
0.001 |
| 24 |
250504_at |
AT5G09840
|
[Putative endonuclease or glycosyl hydrolase] |
0.693 |
-0.013 |
| 25 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.693 |
0.015 |
| 26 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.685 |
0.185 |
| 27 |
260824_at |
AT1G06720
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.680 |
-0.169 |
| 28 |
245879_at |
AT5G09420
|
AtmtOM64, ATTOC64-V, translocon at the outer membrane of chloroplasts 64-V, MTOM64, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-V, OM64, outer membrane 64, TOC64-V, translocon at the outer membrane of chloroplasts 64-V |
0.679 |
-0.032 |
| 29 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
0.662 |
0.051 |
| 30 |
251768_at |
AT3G55940
|
[Phosphoinositide-specific phospholipase C family protein] |
0.661 |
-0.004 |
| 31 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
0.657 |
-0.050 |
| 32 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
0.652 |
0.096 |
| 33 |
246282_at |
AT4G36580
|
[AAA-type ATPase family protein] |
0.644 |
-0.044 |
| 34 |
245807_at |
AT1G46768
|
RAP2.1, related to AP2 1 |
0.638 |
0.029 |
| 35 |
253595_at |
AT4G30830
|
unknown |
0.632 |
-0.038 |
| 36 |
245427_at |
AT4G17550
|
AtG3Pp4, glycerol-3-phosphate permease 4, G3Pp4, glycerol-3-phosphate permease 4 |
0.624 |
0.003 |
| 37 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
0.616 |
0.087 |
| 38 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.610 |
-0.158 |
| 39 |
250418_at |
AT5G11240
|
[transducin family protein / WD-40 repeat family protein] |
0.605 |
-0.142 |
| 40 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
0.594 |
0.004 |
| 41 |
245904_at |
AT5G11110
|
ATSPS2F, sucrose phosphate synthase 2F, KNS2, KAONASHI 2, SPS1, SUCROSE PHOSPHATE SYNTHASE 1, SPS2F, sucrose phosphate synthase 2F, SPSA2, sucrose-phosphate synthase A2 |
0.590 |
0.006 |
| 42 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
0.588 |
-0.101 |
| 43 |
265093_at |
AT1G03905
|
ABCI19, ATP-binding cassette I19 |
0.585 |
0.035 |
| 44 |
248136_at |
AT5G54910
|
[DEA(D/H)-box RNA helicase family protein] |
0.584 |
0.023 |
| 45 |
258965_at |
AT3G10530
|
[Transducin/WD40 repeat-like superfamily protein] |
0.571 |
-0.080 |
| 46 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
0.568 |
-0.027 |
| 47 |
264948_at |
AT1G77030
|
[hydrolases, acting on acid anhydrides, in phosphorus-containing anhydrides] |
0.567 |
-0.098 |
| 48 |
261318_at |
AT1G53035
|
unknown |
0.556 |
-0.060 |
| 49 |
264989_at |
AT1G27200
|
unknown |
0.553 |
-0.022 |
| 50 |
253254_at |
AT4G34650
|
SQS2, squalene synthase 2 |
0.552 |
-0.001 |
| 51 |
245770_at |
AT1G30240
|
[FUNCTIONS IN: binding] |
0.551 |
-0.053 |
| 52 |
257237_at |
AT3G14890
|
[phosphoesterase] |
0.546 |
-0.077 |
| 53 |
262881_at |
AT1G64890
|
[Major facilitator superfamily protein] |
0.545 |
0.030 |
| 54 |
258505_at |
AT3G06530
|
[ARM repeat superfamily protein] |
0.539 |
-0.114 |
| 55 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.535 |
-0.081 |
| 56 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.528 |
-0.215 |
| 57 |
251800_at |
AT3G55510
|
RBL, REBELOTE |
0.528 |
0.003 |
| 58 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
0.525 |
0.063 |
| 59 |
254043_at |
AT4G25990
|
CIL |
0.519 |
-0.043 |
| 60 |
266510_at |
AT2G47990
|
EDA13, EMBRYO SAC DEVELOPMENT ARREST 13, EDA19, EMBRYO SAC DEVELOPMENT ARREST 19, SWA1, SLOW WALKER1 |
0.518 |
-0.053 |
| 61 |
250679_at |
AT5G06550
|
JMJ22, Jumonji domain-containing protein 22 |
0.517 |
-0.118 |
| 62 |
254079_at |
AT4G25730
|
[FtsJ-like methyltransferase family protein] |
0.512 |
-0.223 |
| 63 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.499 |
-0.030 |
| 64 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
0.498 |
-0.236 |
| 65 |
252355_at |
AT3G48250
|
BIR6, Buthionine sulfoximine-insensitive roots 6 |
0.494 |
-0.031 |
| 66 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
0.493 |
0.036 |
| 67 |
257188_at |
AT3G13150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.493 |
0.004 |
| 68 |
265326_at |
AT2G18220
|
[Noc2p family] |
0.492 |
-0.121 |
| 69 |
255225_at |
AT4G05410
|
YAO, YAOZHE |
0.491 |
-0.076 |
| 70 |
260338_at |
AT1G69250
|
[Nuclear transport factor 2 (NTF2) family protein with RNA binding (RRM-RBD-RNP motifs) domain] |
0.489 |
-0.054 |
| 71 |
260585_at |
AT2G43650
|
EMB2777, EMBRYO DEFECTIVE 2777 |
0.487 |
-0.151 |
| 72 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.478 |
-0.071 |
| 73 |
261168_at |
AT1G04945
|
[HIT-type Zinc finger family protein] |
0.478 |
-0.090 |
| 74 |
267309_at |
AT2G19385
|
[zinc ion binding] |
0.478 |
-0.073 |
| 75 |
263574_at |
AT2G16990
|
[Major facilitator superfamily protein] |
0.477 |
0.076 |
| 76 |
250825_at |
AT5G05210
|
[Surfeit locus protein 6] |
0.476 |
-0.090 |
| 77 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.474 |
0.004 |
| 78 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
0.473 |
0.064 |
| 79 |
246484_at |
AT5G16040
|
[Regulator of chromosome condensation (RCC1) family protein] |
0.472 |
-0.082 |
| 80 |
246249_at |
AT4G36680
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.471 |
-0.183 |
| 81 |
263919_at |
AT2G36470
|
unknown |
0.471 |
-0.047 |
| 82 |
250087_at |
AT5G17270
|
[Protein prenylyltransferase superfamily protein] |
0.468 |
0.019 |
| 83 |
247776_at |
AT5G58700
|
ATPLC4, PLC4, phosphatidylinositol-speciwc phospholipase C4 |
0.465 |
-0.027 |
| 84 |
252956_at |
AT4G38580
|
ATFP6, farnesylated protein 6, FP6, farnesylated protein 6, HIPP26, HEAVY METAL ASSOCIATED ISOPRENYLATED PLANT PROTEIN 26 |
0.465 |
0.008 |
| 85 |
265061_at |
AT1G61640
|
[Protein kinase superfamily protein] |
0.464 |
-0.137 |
| 86 |
254076_at |
AT4G25340
|
ATFKBP53, FKBP53, FK506 BINDING PROTEIN 53 |
0.463 |
-0.029 |
| 87 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.462 |
0.017 |
| 88 |
262766_at |
AT1G13160
|
[ARM repeat superfamily protein] |
0.461 |
-0.091 |
| 89 |
256779_at |
AT3G13784
|
AtcwINV5, cell wall invertase 5, CWINV5, cell wall invertase 5 |
0.461 |
0.015 |
| 90 |
251593_at |
AT3G57660
|
NRPA1, nuclear RNA polymerase A1 |
0.461 |
-0.021 |
| 91 |
260331_at |
AT1G80270
|
PPR596, PENTATRICOPEPTIDE REPEAT 596 |
0.461 |
-0.075 |
| 92 |
250009_at |
AT5G18440
|
AtNUFIP, NUFIP, nuclear FMRP-interacting protein |
0.459 |
0.070 |
| 93 |
250379_at |
AT5G11590
|
TINY2, TINY2 |
0.454 |
-0.042 |
| 94 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
0.452 |
0.041 |
| 95 |
261263_at |
AT1G26790
|
[Dof-type zinc finger DNA-binding family protein] |
0.452 |
0.050 |
| 96 |
261377_at |
AT1G18850
|
unknown |
0.452 |
-0.078 |
| 97 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.451 |
-0.080 |
| 98 |
252986_at |
AT4G38380
|
[MATE efflux family protein] |
0.450 |
-0.008 |
| 99 |
257211_at |
AT3G15080
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.450 |
-0.105 |
| 100 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.442 |
-0.035 |
| 101 |
258316_at |
AT3G22660
|
[rRNA processing protein-related] |
0.442 |
-0.102 |
| 102 |
247378_at |
AT5G63120
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.441 |
-0.042 |
| 103 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.439 |
-0.157 |
| 104 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.438 |
-0.026 |
| 105 |
264357_at |
AT1G03360
|
ATRRP4, ribosomal RNA processing 4, RRP4, ribosomal RNA processing 4 |
0.437 |
-0.182 |
| 106 |
253219_at |
AT4G34990
|
AtMYB32, myb domain protein 32, MYB32, myb domain protein 32 |
0.437 |
0.015 |
| 107 |
257694_at |
AT3G12860
|
[NOP56-like pre RNA processing ribonucleoprotein] |
0.436 |
0.018 |
| 108 |
249886_at |
AT5G22320
|
[Leucine-rich repeat (LRR) family protein] |
0.436 |
-0.049 |
| 109 |
267592_at |
AT2G39710
|
[Eukaryotic aspartyl protease family protein] |
0.435 |
-0.050 |
| 110 |
258396_at |
AT3G15460
|
[Ribosomal RNA processing Brix domain protein] |
0.435 |
-0.104 |
| 111 |
247497_at |
AT5G61770
|
PPAN, PETER PAN-like protein |
0.433 |
-0.124 |
| 112 |
259971_at |
AT1G76580
|
[Squamosa promoter-binding protein-like (SBP domain) transcription factor family protein] |
0.430 |
-0.074 |
| 113 |
255866_at |
AT2G30350
|
[Excinuclease ABC, C subunit, N-terminal] |
0.429 |
0.037 |
| 114 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-1.321 |
0.095 |
| 115 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.994 |
0.006 |
| 116 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
-0.971 |
-0.174 |
| 117 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.869 |
0.165 |
| 118 |
256603_at |
AT3G28270
|
unknown |
-0.799 |
-0.101 |
| 119 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
-0.783 |
0.053 |
| 120 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.736 |
0.096 |
| 121 |
252433_at |
AT3G47560
|
[alpha/beta-Hydrolases superfamily protein] |
-0.733 |
0.028 |
| 122 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
-0.680 |
0.013 |
| 123 |
263118_at |
AT1G03090
|
MCCA |
-0.675 |
0.135 |
| 124 |
247977_at |
AT5G56850
|
unknown |
-0.654 |
-0.015 |
| 125 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.651 |
0.193 |
| 126 |
251759_at |
AT3G55630
|
ATDFD, DHFS-FPGS homolog D, DFD, DHFS-FPGS homolog D, FPGS3, folylpolyglutamate synthetase 3 |
-0.648 |
-0.004 |
| 127 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
-0.638 |
0.163 |
| 128 |
256674_at |
AT3G52360
|
unknown |
-0.629 |
-0.024 |
| 129 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.629 |
-0.177 |
| 130 |
247072_at |
AT5G66490
|
unknown |
-0.619 |
-0.001 |
| 131 |
267178_at |
AT2G37750
|
unknown |
-0.608 |
0.016 |
| 132 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.602 |
0.051 |
| 133 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.600 |
-0.061 |
| 134 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
-0.593 |
0.169 |
| 135 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
-0.589 |
-0.039 |
| 136 |
265051_at |
AT1G52100
|
[Mannose-binding lectin superfamily protein] |
-0.586 |
0.015 |
| 137 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
-0.585 |
0.091 |
| 138 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
-0.583 |
0.183 |
| 139 |
260983_at |
AT1G53560
|
[Ribosomal protein L18ae family] |
-0.576 |
0.082 |
| 140 |
250549_at |
AT5G07860
|
[HXXXD-type acyl-transferase family protein] |
-0.575 |
0.013 |
| 141 |
253806_at |
AT4G28270
|
ATRMA2, RMA2, RING membrane-anchor 2 |
-0.565 |
0.113 |
| 142 |
264435_at |
AT1G10360
|
ATGSTU18, glutathione S-transferase TAU 18, GST29, GLUTATHIONE S-TRANSFERASE 29, GSTU18, glutathione S-transferase TAU 18 |
-0.564 |
0.048 |
| 143 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.563 |
0.055 |
| 144 |
260317_at |
AT1G63800
|
UBC5, ubiquitin-conjugating enzyme 5 |
-0.561 |
0.067 |
| 145 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.550 |
0.050 |
| 146 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
-0.548 |
-0.001 |
| 147 |
258275_at |
AT3G15760
|
unknown |
-0.547 |
0.155 |
| 148 |
251369_at |
AT3G60480
|
unknown |
-0.545 |
-0.035 |
| 149 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.532 |
0.159 |
| 150 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.531 |
0.013 |
| 151 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.528 |
0.318 |
| 152 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
-0.520 |
0.100 |
| 153 |
262411_at |
AT1G34640
|
[peptidases] |
-0.515 |
0.056 |
| 154 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-0.506 |
0.038 |
| 155 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
-0.499 |
0.080 |
| 156 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.493 |
0.057 |
| 157 |
255365_at |
AT4G04040
|
MEE51, maternal effect embryo arrest 51 |
-0.471 |
0.005 |
| 158 |
250008_at |
AT5G18630
|
[alpha/beta-Hydrolases superfamily protein] |
-0.471 |
0.145 |
| 159 |
250017_at |
AT5G18140
|
DJC69, DNA J protein A69 |
-0.471 |
0.054 |
| 160 |
257966_at |
AT3G19800
|
unknown |
-0.470 |
0.056 |
| 161 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.470 |
0.087 |
| 162 |
257880_at |
AT3G16910
|
AAE7, acyl-activating enzyme 7, ACN1, ACETATE NON-UTILIZING 1 |
-0.464 |
0.033 |
| 163 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.462 |
0.080 |
| 164 |
258791_at |
AT3G04720
|
AtPR4, HEL, HEVEIN-LIKE, PR-4, PR4, pathogenesis-related 4 |
-0.457 |
0.009 |
| 165 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-0.450 |
0.069 |
| 166 |
247396_at |
AT5G62930
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.450 |
-0.002 |
| 167 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
-0.445 |
0.021 |
| 168 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.443 |
-0.091 |
| 169 |
259545_at |
AT1G20560
|
AAE1, acyl activating enzyme 1 |
-0.442 |
0.031 |
| 170 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.439 |
-0.029 |
| 171 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.425 |
0.080 |
| 172 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.423 |
0.057 |
| 173 |
246462_at |
AT5G16940
|
[carbon-sulfur lyases] |
-0.422 |
0.050 |
| 174 |
260976_at |
AT1G53650
|
CID8, CTC-interacting domain 8 |
-0.422 |
-0.017 |
| 175 |
265149_at |
AT1G51400
|
[Photosystem II 5 kD protein] |
-0.422 |
0.040 |
| 176 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
-0.421 |
0.066 |
| 177 |
266188_at |
AT2G39000
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.420 |
0.046 |
| 178 |
249122_at |
AT5G43850
|
ARD4, ATARD4 |
-0.420 |
0.029 |
| 179 |
253548_at |
AT4G30993
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.418 |
0.049 |
| 180 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
-0.417 |
-0.117 |
| 181 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
-0.415 |
-0.096 |
| 182 |
266106_at |
AT2G45170
|
ATATG8E, AUTOPHAGY 8E, ATG8E, AUTOPHAGY 8E |
-0.414 |
0.119 |
| 183 |
265894_at |
AT2G15050
|
LTP, lipid transfer protein, LTP7, lipid transfer protein 7 |
-0.411 |
0.025 |
| 184 |
254851_at |
AT4G12010
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.410 |
0.103 |
| 185 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
-0.409 |
0.116 |
| 186 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.409 |
0.128 |
| 187 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.405 |
-0.078 |
| 188 |
266984_at |
AT2G39570
|
ACR9, ACT domain repeats 9 |
-0.404 |
0.120 |
| 189 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.404 |
0.105 |
| 190 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.402 |
0.053 |
| 191 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.400 |
0.065 |
| 192 |
247284_at |
AT5G64410
|
ATOPT4, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 4, OPT4, oligopeptide transporter 4 |
-0.399 |
0.109 |
| 193 |
258052_at |
AT3G16190
|
[Isochorismatase family protein] |
-0.399 |
0.027 |
| 194 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.394 |
-0.067 |
| 195 |
246901_at |
AT5G25630
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.390 |
0.058 |
| 196 |
246909_at |
AT5G25770
|
[alpha/beta-Hydrolases superfamily protein] |
-0.386 |
0.027 |
| 197 |
258085_at |
AT3G26100
|
[Regulator of chromosome condensation (RCC1) family protein] |
-0.386 |
0.073 |
| 198 |
258053_at |
AT3G16230
|
[Predicted eukaryotic LigT] |
-0.384 |
-0.015 |
| 199 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.380 |
0.017 |
| 200 |
259708_at |
AT1G77420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.379 |
0.057 |
| 201 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.378 |
0.008 |
| 202 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
-0.377 |
0.010 |
| 203 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.376 |
0.046 |
| 204 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.374 |
0.074 |
| 205 |
261084_at |
AT1G07440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.374 |
0.015 |
| 206 |
263647_at |
AT2G04690
|
Pyridoxamine 5'-phosphate oxidase family protein |
-0.373 |
0.091 |
| 207 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.372 |
-0.057 |
| 208 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
-0.371 |
0.057 |
| 209 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.370 |
0.124 |
| 210 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
-0.368 |
0.124 |
| 211 |
261032_at |
AT1G17430
|
[alpha/beta-Hydrolases superfamily protein] |
-0.367 |
0.007 |
| 212 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.361 |
0.133 |
| 213 |
259061_at |
AT3G07410
|
AtRABA5b, RAB GTPase homolog A5B, RABA5b, RAB GTPase homolog A5B |
-0.360 |
-0.076 |
| 214 |
258375_at |
AT3G17470
|
ATCRSH, CRSH, Ca2+-activated RelA/spot homolog |
-0.360 |
0.075 |
| 215 |
250318_at |
AT5G12200
|
PYD2, pyrimidine 2 |
-0.359 |
0.036 |
| 216 |
251011_at |
AT5G02560
|
HTA12, histone H2A 12 |
-0.359 |
0.014 |
| 217 |
246493_at |
AT5G16180
|
ATCRS1, ARABIDOPSIS ORTHOLOG OF MAIZE CHLOROPLAST SPLICING FACTOR CRS1, CRS1, ortholog of maize chloroplast splicing factor CRS1 |
-0.354 |
0.040 |
| 218 |
266235_at |
AT2G02360
|
AtPP2-B10, phloem protein 2-B10, PP2-B10, phloem protein 2-B10 |
-0.354 |
-0.005 |
| 219 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
-0.351 |
0.053 |
| 220 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
-0.350 |
0.123 |
| 221 |
262749_at |
AT1G28580
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.347 |
0.003 |
| 222 |
257789_at |
AT3G27020
|
AtYSL6, YSL6, YELLOW STRIPE like 6 |
-0.343 |
0.052 |
| 223 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
-0.336 |
0.132 |
| 224 |
247650_at |
AT5G59960
|
unknown |
-0.334 |
0.068 |
| 225 |
261026_at |
AT1G01240
|
unknown |
-0.330 |
0.109 |