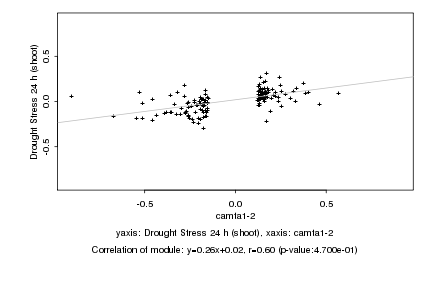
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
camta1-2 |
Log2 signal ratio
Drought Stress 24 h (shoot) |
| 1 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.563 |
0.096 |
| 2 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
0.459 |
-0.025 |
| 3 |
262552_at |
AT1G31350
|
KUF1, KAR-UP F-box 1 |
0.399 |
0.104 |
| 4 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
0.381 |
0.091 |
| 5 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
0.373 |
0.206 |
| 6 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.332 |
0.152 |
| 7 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
0.326 |
0.003 |
| 8 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.316 |
0.111 |
| 9 |
252173_at |
AT3G50650
|
[GRAS family transcription factor] |
0.299 |
0.035 |
| 10 |
252126_at |
AT3G50950
|
ZAR1, HOPZ-ACTIVATED RESISTANCE 1 |
0.271 |
0.079 |
| 11 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
0.253 |
-0.047 |
| 12 |
260312_at |
AT1G63880
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.250 |
0.113 |
| 13 |
253993_at |
AT4G26070
|
ATMEK1, MEK1, MAP kinase/ ERK kinase 1, MKK1, MITOGEN ACTIVATED PROTEIN KINASE KINASE 1, NMAPKK |
0.247 |
0.185 |
| 14 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
0.238 |
0.274 |
| 15 |
266175_at |
AT2G02450
|
ANAC034, Arabidopsis NAC domain containing protein 34, ANAC035, NAC domain containing protein 35, AtLOV1, LOV1, LONG VEGETATIVE PHASE 1, NAC035, NAC domain containing protein 35 |
0.237 |
0.008 |
| 16 |
267550_at |
AT2G32800
|
AP4.3A, LecRK-S.2, L-type lectin receptor kinase S.2 |
0.235 |
0.049 |
| 17 |
258507_at |
AT3G06500
|
A/N-InvC, alkaline/neutral invertase C |
0.218 |
0.106 |
| 18 |
258650_at |
AT3G09830
|
[Protein kinase superfamily protein] |
0.216 |
0.058 |
| 19 |
253827_at |
AT4G28085
|
unknown |
0.206 |
0.072 |
| 20 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
0.202 |
0.143 |
| 21 |
263457_at |
AT2G22300
|
CAMTA3, CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 3, SR1, signal responsive 1 |
0.198 |
0.039 |
| 22 |
249127_at |
AT5G43500
|
ARP9, actin-related protein 9, ATARP9, actin-related protein 9 |
0.192 |
-0.102 |
| 23 |
247940_at |
AT5G57190
|
PSD2, phosphatidylserine decarboxylase 2 |
0.182 |
0.108 |
| 24 |
251684_at |
AT3G56410
|
unknown |
0.178 |
0.127 |
| 25 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
0.174 |
0.153 |
| 26 |
264624_at |
AT1G08930
|
ERD6, EARLY RESPONSE TO DEHYDRATION 6 |
0.172 |
0.052 |
| 27 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.171 |
0.313 |
| 28 |
266019_at |
AT2G18750
|
[Calmodulin-binding protein] |
0.170 |
0.098 |
| 29 |
251109_at |
AT5G01600
|
ATFER1, ARABIDOPSIS THALIANA FERRETIN 1, FER1, ferretin 1 |
0.167 |
-0.219 |
| 30 |
245768_at |
AT1G33590
|
[Leucine-rich repeat (LRR) family protein] |
0.164 |
0.120 |
| 31 |
249284_at |
AT5G41810
|
unknown |
0.164 |
0.083 |
| 32 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.162 |
0.065 |
| 33 |
247126_at |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
0.162 |
0.047 |
| 34 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.161 |
0.065 |
| 35 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
0.161 |
0.228 |
| 36 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
0.159 |
0.109 |
| 37 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.158 |
0.025 |
| 38 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
0.156 |
0.006 |
| 39 |
245765_at |
AT1G33600
|
[Leucine-rich repeat (LRR) family protein] |
0.156 |
0.043 |
| 40 |
254032_at |
AT4G25940
|
[ENTH/ANTH/VHS superfamily protein] |
0.155 |
0.148 |
| 41 |
253414_at |
AT4G33050
|
AtIQM1, EDA39, embryo sac development arrest 39, IQM1, IQ-motif protein 1 |
0.153 |
0.050 |
| 42 |
258282_at |
AT3G26910
|
[hydroxyproline-rich glycoprotein family protein] |
0.153 |
0.023 |
| 43 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
0.151 |
0.212 |
| 44 |
252422_at |
AT3G47550
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.149 |
0.088 |
| 45 |
256567_at |
AT3G19553
|
PUT5, POLYAMINE UPTAKE TRANSPORTER 5 |
0.148 |
0.104 |
| 46 |
267392_at |
AT2G44490
|
BGLU26, BETA GLUCOSIDASE 26, PEN2, PENETRATION 2 |
0.147 |
0.038 |
| 47 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.145 |
0.093 |
| 48 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.144 |
0.135 |
| 49 |
250582_at |
AT5G07580
|
[Integrase-type DNA-binding superfamily protein] |
0.142 |
0.076 |
| 50 |
262711_at |
AT1G16500
|
unknown |
0.139 |
0.064 |
| 51 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.138 |
0.113 |
| 52 |
265245_at |
AT2G43060
|
AtIBH1, IBH1, ILI1 binding bHLH 1 |
0.137 |
0.147 |
| 53 |
263461_at |
AT2G31800
|
[Integrin-linked protein kinase family] |
0.137 |
0.102 |
| 54 |
245050_at |
ATCG00070
|
PSBK, photosystem II reaction center protein K precursor |
0.136 |
0.024 |
| 55 |
260486_at |
AT1G51550
|
[Kelch repeat-containing F-box family protein] |
0.136 |
0.081 |
| 56 |
266069_at |
AT2G18650
|
MEE16, maternal effect embryo arrest 16 |
0.136 |
0.055 |
| 57 |
245642_at |
AT1G25275
|
unknown |
0.136 |
0.081 |
| 58 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
0.134 |
0.069 |
| 59 |
267503_at |
AT2G45600
|
[alpha/beta-Hydrolases superfamily protein] |
0.134 |
0.054 |
| 60 |
256622_at |
AT3G28920
|
AtHB34, homeobox protein 34, HB34, homeobox protein 34, ZHD9, ZINC FINGER HOMEODOMAIN 9 |
0.134 |
0.065 |
| 61 |
252183_at |
AT3G50740
|
UGT72E1, UDP-glucosyl transferase 72E1 |
0.133 |
0.267 |
| 62 |
257516_at |
AT1G69040
|
ACR4, ACT domain repeat 4 |
0.131 |
-0.036 |
| 63 |
261101_at |
AT1G63030
|
ddf2, DWARF AND DELAYED FLOWERING 2 |
0.130 |
-0.018 |
| 64 |
265481_at |
AT2G15960
|
unknown |
0.129 |
0.193 |
| 65 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.129 |
0.128 |
| 66 |
250289_at |
AT5G13190
|
AtGILP, GILP, GSH-induced LITAF domain protein |
0.129 |
0.062 |
| 67 |
260401_at |
AT1G69840
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.127 |
0.038 |
| 68 |
255866_at |
AT2G30350
|
[Excinuclease ABC, C subunit, N-terminal] |
0.126 |
0.015 |
| 69 |
247043_at |
AT5G66880
|
SNRK2-3, SUCROSE NONFERMENTING 1 (SNF1)-RELATED PROTEIN KINASE 2-3, SNRK2.3, sucrose nonfermenting 1(SNF1)-related protein kinase 2.3, SRK2I |
0.125 |
0.114 |
| 70 |
253637_at |
AT4G30390
|
unknown |
0.123 |
0.086 |
| 71 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
0.123 |
-0.037 |
| 72 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
0.122 |
0.167 |
| 73 |
253804_at |
AT4G28230
|
unknown |
0.121 |
0.017 |
| 74 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.907 |
0.064 |
| 75 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.674 |
-0.163 |
| 76 |
246001_at |
AT5G20790
|
unknown |
-0.549 |
-0.184 |
| 77 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.532 |
0.101 |
| 78 |
257421_at |
AT1G12030
|
unknown |
-0.515 |
-0.012 |
| 79 |
256619_at |
AT3G24460
|
[Serinc-domain containing serine and sphingolipid biosynthesis protein] |
-0.515 |
-0.181 |
| 80 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.461 |
-0.202 |
| 81 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-0.461 |
0.025 |
| 82 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.435 |
-0.146 |
| 83 |
252010_at |
AT3G52740
|
unknown |
-0.391 |
-0.125 |
| 84 |
252661_at |
AT3G44450
|
unknown |
-0.385 |
-0.118 |
| 85 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.361 |
0.073 |
| 86 |
264909_at |
AT2G17300
|
unknown |
-0.358 |
-0.121 |
| 87 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.355 |
-0.118 |
| 88 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
-0.341 |
-0.024 |
| 89 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.327 |
-0.136 |
| 90 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.322 |
0.102 |
| 91 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.306 |
-0.142 |
| 92 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.298 |
-0.075 |
| 93 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
-0.285 |
0.060 |
| 94 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.283 |
0.188 |
| 95 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
-0.280 |
-0.119 |
| 96 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
-0.276 |
-0.124 |
| 97 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.271 |
-0.102 |
| 98 |
254683_at |
AT4G13800
|
unknown |
-0.265 |
-0.012 |
| 99 |
266253_at |
AT2G27840
|
HD2D, histone deacetylase 2D, HDA13, HISTONE DEACETYLASE 13, HDT04, HDT4 |
-0.262 |
-0.058 |
| 100 |
254744_at |
AT4G13345
|
MEE55, maternal effect embryo arrest 55 |
-0.261 |
-0.008 |
| 101 |
247055_at |
AT5G66740
|
unknown |
-0.260 |
-0.157 |
| 102 |
250478_at |
AT5G10250
|
DOT3, DEFECTIVELY ORGANIZED TRIBUTARIES 3 |
-0.260 |
-0.146 |
| 103 |
252073_at |
AT3G51750
|
unknown |
-0.255 |
-0.179 |
| 104 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
-0.245 |
-0.049 |
| 105 |
258396_at |
AT3G15460
|
[Ribosomal RNA processing Brix domain protein] |
-0.242 |
-0.193 |
| 106 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.234 |
-0.230 |
| 107 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
-0.231 |
0.013 |
| 108 |
264745_at |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
-0.230 |
-0.002 |
| 109 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
-0.221 |
-0.117 |
| 110 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.220 |
-0.034 |
| 111 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.209 |
-0.041 |
| 112 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.207 |
-0.183 |
| 113 |
251476_at |
AT3G59670
|
unknown |
-0.204 |
-0.233 |
| 114 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.199 |
0.006 |
| 115 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.197 |
-0.083 |
| 116 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.197 |
0.055 |
| 117 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
-0.196 |
-0.013 |
| 118 |
254076_at |
AT4G25340
|
ATFKBP53, FKBP53, FK506 BINDING PROTEIN 53 |
-0.195 |
-0.188 |
| 119 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.189 |
0.023 |
| 120 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
-0.186 |
-0.089 |
| 121 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
-0.185 |
-0.044 |
| 122 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
-0.182 |
0.033 |
| 123 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.179 |
-0.290 |
| 124 |
260331_at |
AT1G80270
|
PPR596, PENTATRICOPEPTIDE REPEAT 596 |
-0.179 |
-0.120 |
| 125 |
267089_at |
AT2G38300
|
[myb-like HTH transcriptional regulator family protein] |
-0.179 |
-0.174 |
| 126 |
252343_at |
AT3G48610
|
NPC6, non-specific phospholipase C6 |
-0.179 |
-0.044 |
| 127 |
251020_at |
AT5G02270
|
ABCI20, ATP-binding cassette I20, ATNAP9, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 9, NAP9, non-intrinsic ABC protein 9 |
-0.176 |
-0.045 |
| 128 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.176 |
0.023 |
| 129 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
-0.176 |
-0.042 |
| 130 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
-0.175 |
-0.009 |
| 131 |
247486_at |
AT5G62140
|
unknown |
-0.174 |
-0.034 |
| 132 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.169 |
0.083 |
| 133 |
253097_at |
AT4G37320
|
CYP81D5, cytochrome P450, family 81, subfamily D, polypeptide 5 |
-0.169 |
-0.159 |
| 134 |
255874_at |
AT2G40550
|
ETG1, E2F target gene 1 |
-0.169 |
-0.094 |
| 135 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.166 |
0.128 |
| 136 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-0.165 |
0.018 |
| 137 |
246765_at |
AT5G27330
|
[Prefoldin chaperone subunit family protein] |
-0.161 |
-0.165 |
| 138 |
265695_at |
AT2G24490
|
ATRPA2, ATRPA32A, ROR1, SUPPRESSOR OF ROS1, RPA2, replicon protein A2, RPA32A |
-0.160 |
-0.118 |
| 139 |
249874_at |
AT5G23070
|
AtTK1b, TK1b, thymidine kinase 1b |
-0.156 |
-0.089 |
| 140 |
260601_at |
AT1G55910
|
ZIP11, zinc transporter 11 precursor |
-0.156 |
-0.003 |
| 141 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.155 |
-0.014 |
| 142 |
248028_at |
AT5G55620
|
unknown |
-0.154 |
-0.070 |
| 143 |
261374_at |
AT1G52990
|
[thioredoxin family protein] |
-0.154 |
0.049 |
| 144 |
255734_at |
AT1G25550
|
[myb-like transcription factor family protein] |
-0.154 |
-0.000 |
| 145 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
-0.153 |
0.038 |