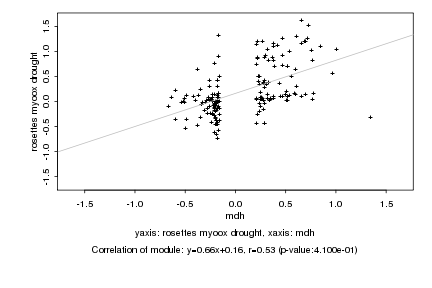
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
mdh |
Log2 signal ratio
rosettes myoox drought |
| 1 |
266393_at |
AT2G41260
|
ATM17, M17 |
1.340 |
-0.313 |
| 2 |
256354_at |
AT1G54870
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.998 |
1.050 |
| 3 |
258327_at |
AT3G22640
|
PAP85 |
0.957 |
0.575 |
| 4 |
263881_at |
AT2G21820
|
unknown |
0.841 |
1.107 |
| 5 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.769 |
0.177 |
| 6 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.761 |
0.831 |
| 7 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
0.759 |
0.050 |
| 8 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
0.753 |
1.039 |
| 9 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
0.726 |
1.535 |
| 10 |
246299_at |
AT3G51810
|
AT3, ATEM1, ARABIDOPSIS THALIANA LATE EMBRYOGENESIS ABUNDANT 1, EM1, LATE EMBRYOGENESIS ABUNDANT 1, GEA1, GUANINE NUCLEOTIDE EXCHANGE FACTOR FOR ADP RIBOSYLATION FACTORS 1 |
0.715 |
1.276 |
| 11 |
262679_at |
AT1G75830
|
LCR67, low-molecular-weight cysteine-rich 67, PDF1.1, PLANT DEFENSIN 1.1 |
0.695 |
0.144 |
| 12 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
0.689 |
1.203 |
| 13 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.686 |
1.202 |
| 14 |
263753_at |
AT2G21490
|
LEA, dehydrin LEA |
0.655 |
1.171 |
| 15 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.653 |
1.639 |
| 16 |
264079_at |
AT2G28490
|
[RmlC-like cupins superfamily protein] |
0.649 |
0.108 |
| 17 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
0.607 |
0.305 |
| 18 |
250624_at |
AT5G07330
|
unknown |
0.600 |
1.302 |
| 19 |
263385_at |
AT2G40170
|
ATEM6, ARABIDOPSIS EARLY METHIONINE-LABELLED 6, EM6, EARLY METHIONINE-LABELLED 6, GEA6, LATE EMBRYOGENESIS ABUNDANT 6 |
0.596 |
0.659 |
| 20 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.593 |
0.157 |
| 21 |
253767_at |
AT4G28520
|
CRC, CRUCIFERIN C, CRU3, cruciferin 3 |
0.583 |
0.171 |
| 22 |
266392_at |
AT2G41280
|
ATM10, M10 |
0.552 |
0.508 |
| 23 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
0.535 |
1.016 |
| 24 |
262828_at |
AT1G14950
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.530 |
0.139 |
| 25 |
264187_at |
AT1G54860
|
[Glycoprotein membrane precursor GPI-anchored] |
0.514 |
0.709 |
| 26 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
0.513 |
0.022 |
| 27 |
254095_at |
AT4G25140
|
OLE1, OLEOSIN 1, OLEO1, oleosin 1 |
0.512 |
0.106 |
| 28 |
245335_at |
AT4G16160
|
ATOEP16-2, ATOEP16-S |
0.510 |
0.702 |
| 29 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
0.501 |
0.204 |
| 30 |
248125_at |
AT5G54740
|
SESA5, seed storage albumin 5 |
0.501 |
0.028 |
| 31 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
0.496 |
0.148 |
| 32 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.468 |
1.262 |
| 33 |
264580_at |
AT1G05340
|
unknown |
0.466 |
0.736 |
| 34 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
0.465 |
0.921 |
| 35 |
265255_at |
AT2G28420
|
GLYI8, glyoxylase I 8 |
0.461 |
0.111 |
| 36 |
251731_at |
AT3G56350
|
[Iron/manganese superoxide dismutase family protein] |
0.440 |
0.107 |
| 37 |
259314_at |
AT3G05260
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.429 |
0.364 |
| 38 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
0.415 |
1.135 |
| 39 |
259167_at |
AT3G01570
|
[Oleosin family protein] |
0.383 |
0.705 |
| 40 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
0.378 |
0.110 |
| 41 |
254203_at |
AT4G24150
|
AtGRF8, growth-regulating factor 8, GRF8, growth-regulating factor 8 |
0.375 |
0.064 |
| 42 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
0.374 |
0.830 |
| 43 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.369 |
1.100 |
| 44 |
252137_at |
AT3G50980
|
XERO1, dehydrin xero 1 |
0.369 |
1.178 |
| 45 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
0.362 |
0.899 |
| 46 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.348 |
0.057 |
| 47 |
250468_at |
AT5G10120
|
[Ethylene insensitive 3 family protein] |
0.344 |
0.063 |
| 48 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.331 |
0.038 |
| 49 |
266654_at |
AT2G25890
|
[Oleosin family protein] |
0.326 |
0.831 |
| 50 |
256938_at |
AT3G22500
|
ATECP31, LATE EMBRYOGENESIS ABUNDANT PROTEIN ECP31 |
0.320 |
0.400 |
| 51 |
263175_at |
AT1G05510
|
OBAP1a, Oil Body-Associated Protein 1a |
0.316 |
1.049 |
| 52 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.315 |
0.075 |
| 53 |
259935_at |
AT1G71250
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.311 |
0.146 |
| 54 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.304 |
0.349 |
| 55 |
249937_at |
AT5G22470
|
[NAD+ ADP-ribosyltransferases] |
0.296 |
0.927 |
| 56 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
0.289 |
-0.430 |
| 57 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.287 |
0.882 |
| 58 |
259478_at |
AT1G18980
|
[RmlC-like cupins superfamily protein] |
0.286 |
-0.036 |
| 59 |
249849_at |
AT5G23230
|
NIC2, nicotinamidase 2 |
0.284 |
0.287 |
| 60 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
0.281 |
-0.421 |
| 61 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.281 |
0.424 |
| 62 |
255007_at |
AT4G10020
|
AtHSD5, hydroxysteroid dehydrogenase 5, HSD5, hydroxysteroid dehydrogenase 5 |
0.279 |
0.032 |
| 63 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.276 |
0.366 |
| 64 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.275 |
-0.146 |
| 65 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.274 |
0.390 |
| 66 |
260088_at |
AT1G73190
|
ALPHA-TIP, ALPHA-TONOPLAST INTRINSIC PROTEIN, TIP3;1 |
0.273 |
0.032 |
| 67 |
249039_at |
AT5G44310
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.265 |
1.212 |
| 68 |
259276_at |
AT3G01190
|
[Peroxidase superfamily protein] |
0.262 |
0.066 |
| 69 |
256815_at |
AT3G21380
|
[Mannose-binding lectin superfamily protein] |
0.257 |
0.045 |
| 70 |
257994_at |
AT3G19920
|
unknown |
0.252 |
0.090 |
| 71 |
254013_at |
AT4G26050
|
PIRL8, plant intracellular ras group-related LRR 8 |
0.248 |
0.183 |
| 72 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.246 |
0.082 |
| 73 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.243 |
-0.083 |
| 74 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
0.241 |
0.060 |
| 75 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.236 |
0.513 |
| 76 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.235 |
0.352 |
| 77 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.235 |
-0.188 |
| 78 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.232 |
-0.032 |
| 79 |
261224_at |
AT1G20160
|
ATSBT5.2 |
0.228 |
0.418 |
| 80 |
256338_at |
AT1G72100
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
0.223 |
0.501 |
| 81 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.223 |
0.508 |
| 82 |
254440_at |
AT4G21020
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.217 |
0.877 |
| 83 |
262260_at |
AT1G70850
|
MLP34, MLP-like protein 34 |
0.217 |
-0.251 |
| 84 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.215 |
1.211 |
| 85 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.211 |
0.899 |
| 86 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.207 |
0.047 |
| 87 |
249013_at |
AT5G44700
|
EDA23, EMBRYO SAC DEVELOPMENT ARREST 23, GSO2, GASSHO 2 |
0.206 |
0.066 |
| 88 |
253870_at |
AT4G27530
|
unknown |
0.205 |
0.744 |
| 89 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.202 |
1.152 |
| 90 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
0.201 |
-0.425 |
| 91 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.671 |
-0.086 |
| 92 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
-0.639 |
0.099 |
| 93 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.600 |
-0.350 |
| 94 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
-0.600 |
0.225 |
| 95 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
-0.538 |
-0.007 |
| 96 |
247024_at |
AT5G66985
|
unknown |
-0.519 |
0.006 |
| 97 |
245226_at |
AT3G29970
|
[B12D protein] |
-0.513 |
-0.018 |
| 98 |
261567_at |
AT1G33055
|
unknown |
-0.508 |
0.079 |
| 99 |
261684_at |
AT1G47400
|
unknown |
-0.507 |
-0.538 |
| 100 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.492 |
-0.354 |
| 101 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.490 |
0.136 |
| 102 |
254200_at |
AT4G24110
|
unknown |
-0.425 |
0.102 |
| 103 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
-0.400 |
0.036 |
| 104 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.382 |
-0.466 |
| 105 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
-0.378 |
0.654 |
| 106 |
257925_at |
AT3G23170
|
unknown |
-0.374 |
0.136 |
| 107 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
-0.356 |
0.251 |
| 108 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
-0.352 |
-0.308 |
| 109 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.347 |
-0.047 |
| 110 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
-0.329 |
0.000 |
| 111 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.311 |
-0.165 |
| 112 |
249384_at |
AT5G39890
|
unknown |
-0.300 |
-0.017 |
| 113 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
-0.298 |
0.023 |
| 114 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
-0.282 |
-0.230 |
| 115 |
250498_at |
AT5G09660
|
PMDH2, peroxisomal NAD-malate dehydrogenase 2 |
-0.281 |
-0.123 |
| 116 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
-0.273 |
0.094 |
| 117 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.271 |
0.053 |
| 118 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-0.267 |
0.433 |
| 119 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
-0.267 |
0.320 |
| 120 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
-0.262 |
0.022 |
| 121 |
251704_at |
AT3G56360
|
unknown |
-0.259 |
-0.088 |
| 122 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.256 |
-0.237 |
| 123 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
-0.249 |
0.042 |
| 124 |
258262_at |
AT3G15770
|
unknown |
-0.245 |
0.048 |
| 125 |
245296_at |
AT4G16370
|
oligopeptide transporter |
-0.245 |
-0.408 |
| 126 |
244980_at |
ATCG00760
|
RPL36, ribosomal protein L36 |
-0.243 |
0.025 |
| 127 |
250152_at |
AT5G15120
|
unknown |
-0.239 |
0.058 |
| 128 |
259685_at |
AT1G63090
|
AtPP2-A11, phloem protein 2-A11, PP2-A11, phloem protein 2-A11 |
-0.236 |
-0.246 |
| 129 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
-0.235 |
0.016 |
| 130 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
-0.229 |
-0.227 |
| 131 |
245163_at |
AT2G33230
|
YUC7, YUCCA 7 |
-0.229 |
0.096 |
| 132 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
-0.229 |
0.148 |
| 133 |
256926_at |
AT3G22540
|
unknown |
-0.225 |
-0.069 |
| 134 |
244965_at |
ATCG00590
|
ORF31 |
-0.218 |
-0.164 |
| 135 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.216 |
-0.611 |
| 136 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
-0.216 |
0.763 |
| 137 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
-0.215 |
-0.033 |
| 138 |
264836_at |
AT1G03610
|
unknown |
-0.215 |
0.153 |
| 139 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
-0.214 |
-0.265 |
| 140 |
245022_at |
ATCG00560
|
PSBL, photosystem II reaction center protein L |
-0.212 |
-0.058 |
| 141 |
244977_at |
ATCG00730
|
PETD, photosynthetic electron transfer D |
-0.211 |
-0.117 |
| 142 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.211 |
-0.131 |
| 143 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.208 |
0.018 |
| 144 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
-0.205 |
-0.311 |
| 145 |
262160_at |
AT1G52590
|
[Putative thiol-disulphide oxidoreductase DCC] |
-0.204 |
-0.072 |
| 146 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.204 |
-0.323 |
| 147 |
256599_at |
AT3G14760
|
unknown |
-0.204 |
-0.444 |
| 148 |
252421_at |
AT3G47540
|
[Chitinase family protein] |
-0.199 |
-0.194 |
| 149 |
244981_at |
ATCG00770
|
RPS8, ribosomal protein S8 |
-0.199 |
-0.074 |
| 150 |
244938_at |
ATCG01120
|
RPS15, chloroplast ribosomal protein S15 |
-0.198 |
-0.373 |
| 151 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.193 |
-0.452 |
| 152 |
245000_at |
ATCG00210
|
YCF6 |
-0.193 |
-0.160 |
| 153 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.192 |
-0.037 |
| 154 |
262170_at |
AT1G74940
|
unknown |
-0.192 |
-0.377 |
| 155 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
-0.190 |
-0.647 |
| 156 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
-0.188 |
-0.130 |
| 157 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
-0.188 |
-0.001 |
| 158 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
-0.187 |
-0.723 |
| 159 |
256262_at |
AT3G12150
|
unknown |
-0.186 |
-0.458 |
| 160 |
255074_at |
AT4G09100
|
[RING/U-box superfamily protein] |
-0.185 |
0.131 |
| 161 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.184 |
0.314 |
| 162 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.183 |
0.081 |
| 163 |
262238_at |
AT1G48300
|
DGAT3, diacylglycerol acyltransferase 3 |
-0.182 |
0.008 |
| 164 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
-0.182 |
0.432 |
| 165 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.182 |
-0.417 |
| 166 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
-0.182 |
0.439 |
| 167 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
-0.179 |
0.024 |
| 168 |
250937_at |
AT5G03230
|
unknown |
-0.177 |
0.161 |
| 169 |
249053_at |
AT5G44440
|
[FAD-binding Berberine family protein] |
-0.173 |
-0.097 |
| 170 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.172 |
-0.575 |
| 171 |
265051_at |
AT1G52100
|
[Mannose-binding lectin superfamily protein] |
-0.171 |
0.176 |
| 172 |
251006_at |
AT5G02600
|
NAKR1, SODIUM POTASSIUM ROOT DEFECTIVE 1, NPCC6, nuclear-enriched phloem companion cell gene 6 |
-0.170 |
0.100 |
| 173 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
-0.170 |
0.901 |
| 174 |
250826_at |
AT5G05220
|
unknown |
-0.169 |
1.338 |
| 175 |
244974_at |
ATCG00700
|
PSBN, photosystem II reaction center protein N |
-0.168 |
0.003 |
| 176 |
258608_at |
AT3G03020
|
unknown |
-0.167 |
-0.258 |
| 177 |
249015_at |
AT5G44730
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.164 |
-0.122 |
| 178 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
-0.163 |
-0.366 |
| 179 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
-0.162 |
0.520 |