|
probeID |
AGICode |
Annotation |
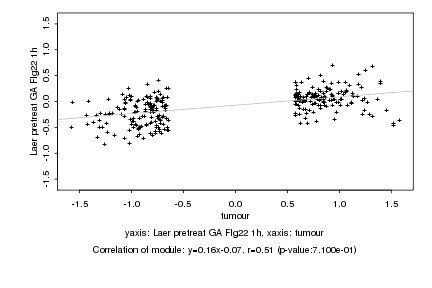
Log2 signal ratio
tumour |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
259009_at |
AT3G09260
|
BGLU23, LEB, LONG ER BODY, PSR3.1, PYK10 |
1.572 |
-0.361 |
| 2 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
1.518 |
-0.450 |
| 3 |
250464_at |
AT5G10040
|
unknown |
1.517 |
-0.423 |
| 4 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
1.448 |
-0.159 |
| 5 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
1.388 |
0.402 |
| 6 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
1.387 |
0.350 |
| 7 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
1.365 |
0.045 |
| 8 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
1.313 |
-0.273 |
| 9 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
1.310 |
0.691 |
| 10 |
257798_at |
AT3G15950
|
NAI2 |
1.281 |
-0.241 |
| 11 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
1.263 |
-0.002 |
| 12 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
1.248 |
0.602 |
| 13 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
1.241 |
0.073 |
| 14 |
257673_at |
AT3G20370
|
[TRAF-like family protein] |
1.234 |
-0.161 |
| 15 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
1.221 |
-0.234 |
| 16 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
1.221 |
0.036 |
| 17 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
1.219 |
0.035 |
| 18 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
1.205 |
0.276 |
| 19 |
260557_at |
AT2G43610
|
[Chitinase family protein] |
1.176 |
0.540 |
| 20 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
1.175 |
0.335 |
| 21 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
1.167 |
0.104 |
| 22 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
1.127 |
0.103 |
| 23 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
1.125 |
0.174 |
| 24 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
1.124 |
0.123 |
| 25 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
1.116 |
-0.017 |
| 26 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
1.110 |
-0.026 |
| 27 |
246855_at |
AT5G26280
|
[TRAF-like family protein] |
1.095 |
0.317 |
| 28 |
252882_at |
AT4G39675
|
unknown |
1.069 |
-0.075 |
| 29 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
1.064 |
0.207 |
| 30 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
1.063 |
0.227 |
| 31 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
1.059 |
0.186 |
| 32 |
245363_at |
AT4G15120
|
[VQ motif-containing protein] |
1.058 |
0.384 |
| 33 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
1.033 |
0.136 |
| 34 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
1.017 |
0.047 |
| 35 |
248254_at |
AT5G53320
|
[Leucine-rich repeat protein kinase family protein] |
1.015 |
0.056 |
| 36 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
1.012 |
0.196 |
| 37 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
1.006 |
0.044 |
| 38 |
254200_at |
AT4G24110
|
unknown |
1.005 |
-0.061 |
| 39 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
0.989 |
-0.005 |
| 40 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.989 |
0.161 |
| 41 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.988 |
0.174 |
| 42 |
259596_at |
AT1G28130
|
GH3.17 |
0.983 |
0.387 |
| 43 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.970 |
-0.208 |
| 44 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.969 |
-0.002 |
| 45 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
0.948 |
-0.333 |
| 46 |
249970_at |
AT5G19100
|
[Eukaryotic aspartyl protease family protein] |
0.948 |
0.044 |
| 47 |
251751_at |
AT3G55720
|
unknown |
0.935 |
0.058 |
| 48 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
0.933 |
-0.069 |
| 49 |
266486_at |
AT2G47950
|
unknown |
0.931 |
0.123 |
| 50 |
251846_at |
AT3G54560
|
HTA11, histone H2A 11 |
0.930 |
0.701 |
| 51 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
0.929 |
0.116 |
| 52 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.923 |
0.036 |
| 53 |
264577_at |
AT1G05260
|
RCI3, RARE COLD INDUCIBLE GENE 3, RCI3A |
0.922 |
0.146 |
| 54 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
0.922 |
0.363 |
| 55 |
267518_at |
AT2G30500
|
NET4B, Networked 4B |
0.910 |
0.241 |
| 56 |
264757_at |
AT1G61360
|
[S-locus lectin protein kinase family protein] |
0.905 |
0.236 |
| 57 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.903 |
0.032 |
| 58 |
248154_at |
AT5G54400
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.883 |
0.260 |
| 59 |
250152_at |
AT5G15120
|
unknown |
0.874 |
0.287 |
| 60 |
258209_at |
AT3G14060
|
unknown |
0.874 |
-0.088 |
| 61 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.871 |
0.211 |
| 62 |
251531_at |
AT3G58550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.858 |
0.084 |
| 63 |
263496_at |
AT2G42570
|
TBL39, TRICHOME BIREFRINGENCE-LIKE 39 |
0.857 |
-0.073 |
| 64 |
245208_at |
AT5G12330
|
LRP1, LATERAL ROOT PRIMORDIUM 1 |
0.854 |
0.110 |
| 65 |
260528_at |
AT2G47260
|
ATWRKY23, WRKY DNA-BINDING PROTEIN 23, WRKY23, WRKY DNA-binding protein 23 |
0.851 |
-0.007 |
| 66 |
250070_at |
AT5G17980
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.849 |
-0.073 |
| 67 |
247625_at |
AT5G60200
|
TMO6, TARGET OF MONOPTEROS 6 |
0.847 |
0.010 |
| 68 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.843 |
0.393 |
| 69 |
257952_at |
AT3G21770
|
[Peroxidase superfamily protein] |
0.843 |
-0.016 |
| 70 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.842 |
0.156 |
| 71 |
246933_at |
AT5G25160
|
ZFP3, zinc finger protein 3 |
0.840 |
0.052 |
| 72 |
260234_at |
AT1G74460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.840 |
0.055 |
| 73 |
254898_at |
AT4G11480
|
CRK32, cysteine-rich RLK (RECEPTOR-like protein kinase) 32 |
0.823 |
0.089 |
| 74 |
260181_at |
AT1G70710
|
ATGH9B1, glycosyl hydrolase 9B1, CEL1, CELLULASE 1, GH9B1, glycosyl hydrolase 9B1 |
0.817 |
-0.117 |
| 75 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.817 |
0.521 |
| 76 |
252751_at |
AT3G43430
|
[RING/U-box superfamily protein] |
0.816 |
0.201 |
| 77 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
0.815 |
-0.067 |
| 78 |
245328_at |
AT4G14465
|
AHL20, AT-hook motif nuclear-localized protein 20 |
0.810 |
0.195 |
| 79 |
262260_at |
AT1G70850
|
MLP34, MLP-like protein 34 |
0.806 |
0.038 |
| 80 |
266469_at |
AT2G31180
|
ATMYB14, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 14, MYB14, myb domain protein 14, MYB14AT |
0.804 |
0.052 |
| 81 |
261567_at |
AT1G33055
|
unknown |
0.795 |
-0.069 |
| 82 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
0.794 |
0.239 |
| 83 |
254608_at |
AT4G18910
|
ATNLM2, NOD26-LIKE INTRINSIC PROTEIN 2, NIP1;2, NOD26-like intrinsic protein 1;2, NLM2, NOD26-LIKE INTRINSIC PROTEIN 2 |
0.794 |
0.124 |
| 84 |
258618_at |
AT3G02885
|
GASA5, GAST1 protein homolog 5 |
0.783 |
0.046 |
| 85 |
247069_at |
AT5G66920
|
sks17, SKU5 similar 17 |
0.774 |
-0.374 |
| 86 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
0.771 |
0.112 |
| 87 |
262813_at |
AT1G11670
|
[MATE efflux family protein] |
0.770 |
-0.037 |
| 88 |
263438_at |
AT2G28660
|
[Chloroplast-targeted copper chaperone protein] |
0.768 |
0.165 |
| 89 |
261585_at |
AT1G01010
|
ANAC001, NAC domain containing protein 1, NAC001, NAC domain containing protein 1 |
0.756 |
0.134 |
| 90 |
254754_at |
AT4G13210
|
[Pectin lyase-like superfamily protein] |
0.754 |
0.080 |
| 91 |
260435_at |
AT1G68320
|
AtMYB62, myb domain protein 62, BW62B, BW62C, MYB62, myb domain protein 62 |
0.753 |
0.120 |
| 92 |
260333_at |
AT1G70500
|
[Pectin lyase-like superfamily protein] |
0.752 |
-0.034 |
| 93 |
261460_at |
AT1G07880
|
ATMPK13 |
0.749 |
0.078 |
| 94 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.746 |
0.099 |
| 95 |
256681_at |
AT3G52340
|
ATSPP2, SUCROSE-PHOSPHATASE 2, SPP2, sucrose-6F-phosphate phosphohydrolase 2 |
0.745 |
-0.206 |
| 96 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
0.742 |
0.086 |
| 97 |
264815_at |
AT1G03620
|
[ELMO/CED-12 family protein] |
0.737 |
0.109 |
| 98 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
0.736 |
0.143 |
| 99 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.735 |
0.279 |
| 100 |
263194_at |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
0.735 |
-0.048 |
| 101 |
249384_at |
AT5G39890
|
unknown |
0.729 |
0.082 |
| 102 |
254756_at |
AT4G13235
|
EDA21, embryo sac development arrest 21 |
0.718 |
0.002 |
| 103 |
247492_at |
AT5G61890
|
[Integrase-type DNA-binding superfamily protein] |
0.707 |
0.162 |
| 104 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
0.704 |
0.180 |
| 105 |
264001_at |
AT2G22420
|
[Peroxidase superfamily protein] |
0.704 |
-0.171 |
| 106 |
251612_at |
AT3G57950
|
unknown |
0.694 |
0.454 |
| 107 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.692 |
0.142 |
| 108 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
0.690 |
0.155 |
| 109 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
0.687 |
-0.413 |
| 110 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
0.680 |
0.042 |
| 111 |
263998_at |
AT2G22510
|
[hydroxyproline-rich glycoprotein family protein] |
0.679 |
0.098 |
| 112 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.676 |
-0.213 |
| 113 |
262913_at |
AT1G59960
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.674 |
-0.309 |
| 114 |
261518_at |
AT1G71695
|
[Peroxidase superfamily protein] |
0.673 |
-0.137 |
| 115 |
250756_at |
AT5G05940
|
ATROPGEF5, ROP GUANINE NUCLEOTIDE EXCHANGE FACTOR 5, ROPGEF5, ROP (rho of plants) guanine nucleotide exchange factor 5 |
0.665 |
0.018 |
| 116 |
251858_at |
AT3G54820
|
PIP2;5, plasma membrane intrinsic protein 2;5, PIP2D, PLASMA MEMBRANE INTRINSIC PROTEIN 2D |
0.653 |
-0.141 |
| 117 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
0.650 |
0.086 |
| 118 |
250753_at |
AT5G05860
|
UGT76C2, UDP-glucosyl transferase 76C2 |
0.647 |
0.022 |
| 119 |
250426_at |
AT5G10510
|
AIL6, AINTEGUMENTA-like 6, PLT3, PLETHORA 3 |
0.646 |
0.056 |
| 120 |
247196_at |
AT5G65510
|
AIL7, AINTEGUMENTA-like 7, PLT7, PLETHORA 7 |
0.644 |
-0.006 |
| 121 |
249060_at |
AT5G44560
|
VPS2.2 |
0.644 |
-0.017 |
| 122 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.633 |
0.377 |
| 123 |
246093_at |
AT5G20550
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.624 |
0.042 |
| 124 |
247904_at |
AT5G57390
|
AIL5, AINTEGUMENTA-like 5, CHO1, CHOTTO 1, EMK, EMBRYOMAKER, PLT5, PLETHORA 5 |
0.624 |
0.049 |
| 125 |
253493_at |
AT4G31820
|
ENP, ENHANCER OF PINOID, MAB4, MACCHI-BOU 4, NPY1, NAKED PINS IN YUC MUTANTS 1 |
0.622 |
-0.411 |
| 126 |
260395_at |
AT1G69780
|
ATHB13 |
0.618 |
0.099 |
| 127 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
0.615 |
-0.246 |
| 128 |
264176_at |
AT1G02110
|
unknown |
0.613 |
-0.120 |
| 129 |
258623_at |
AT3G02790
|
MBS1, METHYLENE BLUE SENSITIVITY 1 |
0.611 |
0.050 |
| 130 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
0.610 |
0.193 |
| 131 |
260053_at |
AT1G78120
|
TPR12, tetratricopeptide repeat 12 |
0.609 |
0.009 |
| 132 |
248420_at |
AT5G51560
|
[Leucine-rich repeat protein kinase family protein] |
0.605 |
-0.065 |
| 133 |
249824_at |
AT5G23380
|
unknown |
0.604 |
0.063 |
| 134 |
255782_at |
AT1G19850
|
ARF5, AUXIN RESPONSE FACTOR 5, IAA24, indole-3-acetic acid inducible 24, MP, MONOPTEROS |
0.604 |
0.147 |
| 135 |
259599_at |
AT1G28110
|
SCPL45, serine carboxypeptidase-like 45 |
0.594 |
0.020 |
| 136 |
253828_at |
AT4G27970
|
SLAH2, SLAC1 homologue 2 |
0.592 |
0.103 |
| 137 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.591 |
0.144 |
| 138 |
255596_at |
AT4G01720
|
AtWRKY47, WRKY47 |
0.587 |
0.116 |
| 139 |
247149_at |
AT5G65660
|
[hydroxyproline-rich glycoprotein family protein] |
0.584 |
-0.055 |
| 140 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
0.582 |
0.163 |
| 141 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.581 |
0.250 |
| 142 |
260392_at |
AT1G74030
|
ENO1, enolase 1 |
0.581 |
0.322 |
| 143 |
263734_at |
AT1G60030
|
ATNAT7, ARABIDOPSIS NUCLEOBASE-ASCORBATE TRANSPORTER 7, NAT7, nucleobase-ascorbate transporter 7 |
0.579 |
0.113 |
| 144 |
260555_at |
AT2G41780
|
unknown |
0.579 |
0.098 |
| 145 |
250180_at |
AT5G14450
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.577 |
-0.044 |
| 146 |
256839_at |
AT3G22930
|
AtCML11, CML11, calmodulin-like 11 |
0.577 |
-0.222 |
| 147 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
0.577 |
0.016 |
| 148 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.575 |
0.368 |
| 149 |
247359_at |
AT5G63560
|
FACT, FATTY ALCOHOL:CAFFEOYL-CoA CAFFEOYL TRANSFERASE |
0.574 |
0.083 |
| 150 |
262942_at |
AT1G79450
|
ALIS5, ALA-interacting subunit 5 |
0.572 |
0.372 |
| 151 |
267624_at |
AT2G39660
|
BIK1, botrytis-induced kinase1 |
0.571 |
0.132 |
| 152 |
262802_at |
AT1G20930
|
CDKB2;2, cyclin-dependent kinase B2;2 |
0.570 |
0.141 |
| 153 |
264790_at |
AT2G17820
|
AHK1, ATHK1, histidine kinase 1, HK1, histidine kinase 1 |
0.570 |
-0.256 |
| 154 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-1.585 |
-0.494 |
| 155 |
267262_at |
AT2G22990
|
SCPL8, SERINE CARBOXYPEPTIDASE-LIKE 8, SNG1, sinapoylglucose 1 |
-1.570 |
-0.004 |
| 156 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-1.443 |
-0.259 |
| 157 |
256603_at |
AT3G28270
|
unknown |
-1.426 |
-0.432 |
| 158 |
260007_at |
AT1G67870
|
[glycine-rich protein] |
-1.415 |
0.003 |
| 159 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-1.374 |
-0.395 |
| 160 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-1.343 |
-0.259 |
| 161 |
263709_at |
AT1G09310
|
unknown |
-1.332 |
-0.678 |
| 162 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-1.312 |
-0.488 |
| 163 |
252092_at |
AT3G51420
|
ATSSL4, STRICTOSIDINE SYNTHASE-LIKE 4, SSL4, strictosidine synthase-like 4 |
-1.309 |
-0.354 |
| 164 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-1.302 |
-0.217 |
| 165 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-1.286 |
-0.484 |
| 166 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-1.265 |
-0.814 |
| 167 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-1.253 |
-0.241 |
| 168 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-1.250 |
-0.414 |
| 169 |
248028_at |
AT5G55620
|
unknown |
-1.233 |
-0.590 |
| 170 |
258091_at |
AT3G14560
|
unknown |
-1.226 |
0.052 |
| 171 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-1.220 |
-0.236 |
| 172 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
-1.216 |
-0.218 |
| 173 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-1.193 |
-0.228 |
| 174 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
-1.170 |
-0.642 |
| 175 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
-1.137 |
-0.103 |
| 176 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-1.123 |
-0.142 |
| 177 |
250498_at |
AT5G09660
|
PMDH2, peroxisomal NAD-malate dehydrogenase 2 |
-1.099 |
-0.246 |
| 178 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
-1.089 |
0.144 |
| 179 |
262811_at |
AT1G11700
|
unknown |
-1.076 |
-0.121 |
| 180 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-1.075 |
-0.698 |
| 181 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
-1.075 |
-0.277 |
| 182 |
258746_at |
AT3G05950
|
[RmlC-like cupins superfamily protein] |
-1.071 |
0.046 |
| 183 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-1.068 |
-0.150 |
| 184 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-1.065 |
-0.002 |
| 185 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-1.059 |
-0.032 |
| 186 |
259766_at |
AT1G64360
|
unknown |
-1.051 |
0.090 |
| 187 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-1.032 |
0.268 |
| 188 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-1.028 |
0.118 |
| 189 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
-1.022 |
-0.811 |
| 190 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
-1.021 |
-0.559 |
| 191 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-1.018 |
0.027 |
| 192 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-1.013 |
-0.329 |
| 193 |
257872_at |
AT3G28360
|
ABCB16, ATP-binding cassette B16, PGP16, P-glycoprotein 16 |
-1.012 |
0.042 |
| 194 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
-1.009 |
0.116 |
| 195 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-1.005 |
-0.436 |
| 196 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
-0.997 |
-0.375 |
| 197 |
263410_at |
AT2G04039
|
unknown |
-0.986 |
-0.311 |
| 198 |
245701_at |
AT5G04140
|
FD-GOGAT, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE, GLS1, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1, GLU1, glutamate synthase 1, GLUS |
-0.985 |
-0.378 |
| 199 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
-0.984 |
-0.483 |
| 200 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.984 |
-0.371 |
| 201 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.974 |
-0.200 |
| 202 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.964 |
-0.443 |
| 203 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.964 |
-0.077 |
| 204 |
263350_at |
AT2G13360
|
AGT, alanine:glyoxylate aminotransferase, AGT1, ALANINE:GLYOXYLATE AMINOTRANSFERASE 1, SGAT, L-serine:glyoxylate aminotransferase |
-0.963 |
-0.060 |
| 205 |
265305_at |
AT2G20340
|
AAS, aromatic aldehyde synthase, AtAAS |
-0.963 |
-0.193 |
| 206 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.961 |
-0.461 |
| 207 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
-0.954 |
-0.107 |
| 208 |
257509_at |
AT1G63190
|
[Cystatin/monellin superfamily protein] |
-0.952 |
0.018 |
| 209 |
261581_at |
AT1G01140
|
CIPK9, CBL-interacting protein kinase 9, PKS6, PROTEIN KINASE 6, SnRK3.12, SNF1-RELATED PROTEIN KINASE 3.12 |
-0.947 |
-0.181 |
| 210 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
-0.946 |
-0.658 |
| 211 |
265025_at |
AT1G24575
|
unknown |
-0.940 |
0.028 |
| 212 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.938 |
-0.538 |
| 213 |
248153_at |
AT5G54250
|
ATCNGC4, cyclic nucleotide-gated cation channel 4, CNGC4, cyclic nucleotide-gated cation channel 4, DND2, DEFENSE, NO DEATH 2, HLM1 |
-0.934 |
-0.518 |
| 214 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.932 |
-0.486 |
| 215 |
256870_at |
AT3G26300
|
CYP71B34, cytochrome P450, family 71, subfamily B, polypeptide 34 |
-0.928 |
-0.622 |
| 216 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
-0.927 |
-0.319 |
| 217 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.921 |
-0.488 |
| 218 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.905 |
-0.470 |
| 219 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
-0.903 |
-0.272 |
| 220 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.892 |
-0.714 |
| 221 |
248218_at |
AT5G53710
|
unknown |
-0.891 |
0.054 |
| 222 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
-0.876 |
-0.272 |
| 223 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.875 |
-0.042 |
| 224 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
-0.870 |
-0.190 |
| 225 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.867 |
-0.142 |
| 226 |
250624_at |
AT5G07330
|
unknown |
-0.863 |
0.103 |
| 227 |
248329_at |
AT5G52780
|
unknown |
-0.857 |
-0.450 |
| 228 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.857 |
-0.442 |
| 229 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
-0.854 |
0.092 |
| 230 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.848 |
0.345 |
| 231 |
259765_at |
AT1G64370
|
unknown |
-0.848 |
-0.008 |
| 232 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.837 |
-0.053 |
| 233 |
250485_at |
AT5G09990
|
PROPEP5, elicitor peptide 5 precursor |
-0.827 |
-0.208 |
| 234 |
256542_at |
AT1G42550
|
PMI1, PLASTID MOVEMENT IMPAIRED1 |
-0.827 |
-0.152 |
| 235 |
266979_at |
AT2G39470
|
PnsL1, Photosynthetic NDH subcomplex L 1, PPL2, PsbP-like protein 2 |
-0.825 |
-0.600 |
| 236 |
261788_at |
AT1G15980
|
NDF1, NDH-dependent cyclic electron flow 1, NDH48, NAD(P)H DEHYDROGENASE SUBUNIT 48, PnsB1, Photosynthetic NDH subcomplex B 1 |
-0.825 |
-0.415 |
| 237 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.825 |
0.098 |
| 238 |
260851_at |
AT1G21890
|
UMAMIT19, Usually multiple acids move in and out Transporters 19 |
-0.822 |
-0.019 |
| 239 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
-0.819 |
-0.083 |
| 240 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
-0.819 |
-0.103 |
| 241 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
-0.818 |
0.070 |
| 242 |
252876_at |
AT4G39970
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.817 |
-0.066 |
| 243 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.815 |
-0.115 |
| 244 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
-0.810 |
-0.391 |
| 245 |
256754_at |
AT3G25690
|
AtCHUP1, Arabidopsis thaliana CHLOROPLAST UNUSUAL POSITIONING 1, CHUP1, CHLOROPLAST UNUSUAL POSITIONING 1 |
-0.808 |
-0.293 |
| 246 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
-0.805 |
-0.103 |
| 247 |
256145_at |
AT1G48750
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.804 |
-0.164 |
| 248 |
252366_at |
AT3G48420
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.802 |
-0.643 |
| 249 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.798 |
-0.196 |
| 250 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.797 |
-0.155 |
| 251 |
249136_at |
AT5G43180
|
unknown |
-0.792 |
-0.066 |
| 252 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.790 |
-0.172 |
| 253 |
251986_at |
AT3G53310
|
[AP2/B3-like transcriptional factor family protein] |
-0.781 |
-0.068 |
| 254 |
251906_at |
AT3G53720
|
ATCHX20, cation/H+ exchanger 20, CHX20, cation/H+ exchanger 20 |
-0.780 |
0.176 |
| 255 |
255889_at |
AT1G17840
|
ABCG11, ATP-binding cassette G11, AtABCG11, ATWBC11, ARABIDOPSIS THALIANA WHITE-BROWN COMPLEX HOMOLOG PROTEIN 11, COF1, CUTICULAR DEFECT AND ORGAN FUSION 1, DSO, DESPERADO, WBC11, white-brown complex homolog protein 11 |
-0.778 |
-0.608 |
| 256 |
261224_at |
AT1G20160
|
ATSBT5.2 |
-0.775 |
-0.369 |
| 257 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.775 |
-0.575 |
| 258 |
267264_at |
AT2G22970
|
SCPL11, serine carboxypeptidase-like 11 |
-0.768 |
-0.161 |
| 259 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.765 |
-0.460 |
| 260 |
247713_at |
AT5G59330
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.764 |
0.088 |
| 261 |
260060_at |
AT1G73680
|
ALPHA DOX2, alpha dioxygenase, alpha-DOX2, alpha-dioxygenase 2 |
-0.762 |
-0.118 |
| 262 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
-0.761 |
0.007 |
| 263 |
256015_at |
AT1G19150
|
LHCA2*1, Lhca6, photosystem I light harvesting complex gene 6 |
-0.761 |
-0.502 |
| 264 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.759 |
0.074 |
| 265 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.758 |
0.040 |
| 266 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.757 |
0.036 |
| 267 |
254153_at |
AT4G24450
|
ATGWD2, GWD3, PWD, phosphoglucan, water dikinase |
-0.752 |
-0.279 |
| 268 |
245243_at |
AT1G44414
|
unknown |
-0.750 |
0.197 |
| 269 |
260149_at |
AT1G52810
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.748 |
0.411 |
| 270 |
252353_at |
AT3G48200
|
unknown |
-0.742 |
-0.501 |
| 271 |
263947_at |
AT2G35820
|
[ureidoglycolate hydrolases] |
-0.742 |
-0.319 |
| 272 |
254122_at |
AT4G24510
|
CER2, ECERIFERUM 2, VC-2, VC2 |
-0.739 |
-0.330 |
| 273 |
249120_at |
AT5G43750
|
NDH18, NAD(P)H dehydrogenase 18, PnsB5, Photosynthetic NDH subcomplex B 5 |
-0.738 |
-0.578 |
| 274 |
257954_at |
AT3G21760
|
HYR1, HYPOSTATIN RESISTANCE 1 |
-0.737 |
-0.533 |
| 275 |
255719_at |
AT1G32080
|
AtLrgB, LrgB |
-0.735 |
-0.267 |
| 276 |
252327_at |
AT3G48740
|
AtSWEET11, SWEET11 |
-0.727 |
0.009 |
| 277 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.727 |
-0.356 |
| 278 |
262693_at |
AT1G62780
|
unknown |
-0.726 |
-0.422 |
| 279 |
253689_at |
AT4G29770
|
unknown |
-0.724 |
0.145 |
| 280 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.714 |
-0.616 |
| 281 |
267036_at |
AT2G38465
|
unknown |
-0.711 |
-0.090 |
| 282 |
251591_at |
AT3G57680
|
[Peptidase S41 family protein] |
-0.709 |
-0.016 |
| 283 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.706 |
0.065 |
| 284 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
-0.704 |
0.030 |
| 285 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.703 |
-0.298 |
| 286 |
261084_at |
AT1G07440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.701 |
-0.430 |
| 287 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.698 |
-0.278 |
| 288 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.695 |
-0.283 |
| 289 |
246342_at |
AT3G56700
|
AtFAR6, FAR6, fatty acid reductase 6 |
-0.695 |
0.083 |
| 290 |
262916_at |
AT1G59700
|
ATGSTU16, glutathione S-transferase TAU 16, GSTU16, glutathione S-transferase TAU 16 |
-0.691 |
-0.526 |
| 291 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
-0.687 |
-0.084 |
| 292 |
266516_at |
AT2G47880
|
[Glutaredoxin family protein] |
-0.685 |
-0.086 |
| 293 |
247357_at |
AT5G63710
|
[Leucine-rich repeat protein kinase family protein] |
-0.681 |
-0.157 |
| 294 |
254174_at |
AT4G24120
|
ATYSL1, YELLOW STRIPE LIKE 1, YSL1, YELLOW STRIPE like 1 |
-0.680 |
-0.131 |
| 295 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.677 |
-0.090 |
| 296 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.675 |
-0.067 |
| 297 |
251987_at |
AT3G53280
|
CYP71B5, cytochrome p450 71b5 |
-0.673 |
0.260 |
| 298 |
262728_at |
AT1G75820
|
ATCLV1, CLV1, CLAVATA 1, FAS3, FASCIATA 3, FLO5, FLOWER DEVELOPMENT 5 |
-0.670 |
-0.322 |
| 299 |
260076_at |
AT1G73630
|
[EF hand calcium-binding protein family] |
-0.668 |
-0.069 |
| 300 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.661 |
-0.551 |
| 301 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.659 |
0.094 |
| 302 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.656 |
-0.498 |
| 303 |
245876_at |
AT1G26230
|
Cpn60beta4, chaperonin-60beta4 |
-0.651 |
-0.544 |
| 304 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
-0.650 |
0.268 |
| 305 |
250960_at |
AT5G02940
|
unknown |
-0.645 |
-0.351 |