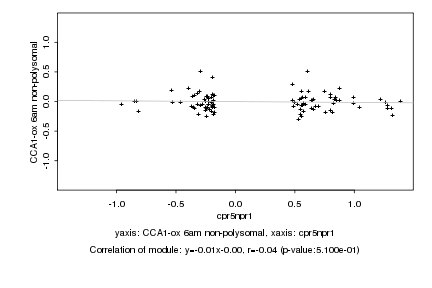
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
cpr5npr1 |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
1.385 |
0.011 |
| 2 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
1.313 |
-0.234 |
| 3 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.308 |
-0.109 |
| 4 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
1.275 |
-0.052 |
| 5 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
1.272 |
-0.112 |
| 6 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
1.256 |
-0.016 |
| 7 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
1.216 |
0.042 |
| 8 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
1.037 |
-0.097 |
| 9 |
261545_at |
AT1G63530
|
unknown |
0.991 |
-0.032 |
| 10 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.988 |
0.082 |
| 11 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.875 |
0.235 |
| 12 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.867 |
0.025 |
| 13 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.846 |
0.021 |
| 14 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.834 |
0.078 |
| 15 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
0.832 |
0.039 |
| 16 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.819 |
-0.027 |
| 17 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.810 |
-0.175 |
| 18 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.799 |
0.128 |
| 19 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.796 |
0.074 |
| 20 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.792 |
-0.146 |
| 21 |
266017_at |
AT2G18690
|
unknown |
0.755 |
-0.176 |
| 22 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.741 |
0.180 |
| 23 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
0.695 |
-0.081 |
| 24 |
264580_at |
AT1G05340
|
unknown |
0.665 |
-0.073 |
| 25 |
249454_at |
AT5G39520
|
unknown |
0.656 |
0.035 |
| 26 |
264635_at |
AT1G65500
|
unknown |
0.652 |
-0.118 |
| 27 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.647 |
0.028 |
| 28 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.635 |
-0.111 |
| 29 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
0.633 |
0.023 |
| 30 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.608 |
0.176 |
| 31 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
0.606 |
0.509 |
| 32 |
256050_at |
AT1G07000
|
ATEXO70B2, exocyst subunit exo70 family protein B2, EXO70B2, exocyst subunit exo70 family protein B2 |
0.589 |
0.070 |
| 33 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
0.583 |
-0.036 |
| 34 |
255148_at |
AT4G08470
|
MAPKKK10, MEKK3, MAPK/ERK kinase kinase 3 |
0.567 |
-0.030 |
| 35 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.565 |
-0.156 |
| 36 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.563 |
-0.060 |
| 37 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.556 |
0.072 |
| 38 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.554 |
0.179 |
| 39 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.553 |
-0.051 |
| 40 |
267178_at |
AT2G37750
|
unknown |
0.551 |
-0.248 |
| 41 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
0.548 |
0.059 |
| 42 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.547 |
-0.118 |
| 43 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
0.541 |
-0.216 |
| 44 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.531 |
0.041 |
| 45 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.523 |
-0.300 |
| 46 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.516 |
-0.034 |
| 47 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
0.495 |
-0.002 |
| 48 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
0.487 |
-0.073 |
| 49 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.475 |
0.028 |
| 50 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
0.475 |
0.301 |
| 51 |
261258_at |
AT1G26640
|
[Amino acid kinase family protein] |
-0.965 |
-0.046 |
| 52 |
261273_at |
AT1G26650
|
unknown |
-0.853 |
0.012 |
| 53 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.833 |
0.015 |
| 54 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.818 |
-0.157 |
| 55 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.540 |
0.187 |
| 56 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
-0.532 |
-0.006 |
| 57 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.466 |
-0.009 |
| 58 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
-0.396 |
0.231 |
| 59 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.373 |
-0.078 |
| 60 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.369 |
0.095 |
| 61 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.354 |
-0.086 |
| 62 |
264909_at |
AT2G17300
|
unknown |
-0.352 |
0.109 |
| 63 |
253966_at |
AT4G26520
|
AtFBA7, FBA7, fructose-bisphosphate aldolase 7 |
-0.345 |
-0.103 |
| 64 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.326 |
0.142 |
| 65 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.322 |
-0.038 |
| 66 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.317 |
-0.214 |
| 67 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.309 |
0.186 |
| 68 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.300 |
0.510 |
| 69 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.299 |
-0.058 |
| 70 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
-0.282 |
-0.034 |
| 71 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
-0.263 |
0.036 |
| 72 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.260 |
0.012 |
| 73 |
261597_at |
AT1G49780
|
PUB26, plant U-box 26 |
-0.258 |
-0.143 |
| 74 |
253814_at |
AT4G28290
|
unknown |
-0.257 |
-0.099 |
| 75 |
263788_at |
AT2G24580
|
[FAD-dependent oxidoreductase family protein] |
-0.250 |
-0.243 |
| 76 |
250724_at |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.249 |
0.093 |
| 77 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.246 |
-0.117 |
| 78 |
259346_at |
AT3G03910
|
glutamate dehydrogenase 3 |
-0.241 |
-0.100 |
| 79 |
267545_at |
AT2G32690
|
ATGRP23, GLYCINE-RICH PROTEIN 23, GRP23, glycine-rich protein 23 |
-0.239 |
0.101 |
| 80 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
-0.234 |
-0.041 |
| 81 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.232 |
0.054 |
| 82 |
246915_at |
AT5G25880
|
ATNADP-ME3, Arabidopsis thaliana NADP-malic enzyme 3, NADP-ME3, NADP-malic enzyme 3 |
-0.232 |
0.008 |
| 83 |
250696_at |
AT5G06790
|
unknown |
-0.227 |
-0.034 |
| 84 |
267359_at |
AT2G40020
|
[Nucleolar histone methyltransferase-related protein] |
-0.224 |
-0.119 |
| 85 |
265387_at |
AT2G20670
|
unknown |
-0.208 |
0.083 |
| 86 |
246540_at |
AT5G15600
|
SP1L4, SPIRAL1-like4 |
-0.206 |
-0.152 |
| 87 |
258782_at |
AT3G11750
|
FOLB1 |
-0.204 |
-0.012 |
| 88 |
247118_at |
AT5G65890
|
ACR1, ACT domain repeat 1 |
-0.198 |
-0.082 |
| 89 |
249510_at |
AT5G38510
|
[Rhomboid-related intramembrane serine protease family protein] |
-0.198 |
0.418 |
| 90 |
266529_at |
AT2G16880
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.195 |
-0.087 |
| 91 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
-0.194 |
0.119 |
| 92 |
264378_at |
AT2G25220
|
[Protein kinase superfamily protein] |
-0.193 |
-0.062 |
| 93 |
247940_at |
AT5G57190
|
PSD2, phosphatidylserine decarboxylase 2 |
-0.191 |
0.023 |
| 94 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.189 |
-0.219 |
| 95 |
258034_at |
AT3G21300
|
[RNA methyltransferase family protein] |
-0.187 |
-0.050 |
| 96 |
246330_at |
AT3G43600
|
AAO2, aldehyde oxidase 2, AO3, aldehyde oxidase 3, AOgamma, Aldehyde oxidase gamma, atAO-2, Arabidopsis thaliana aldehyde oxidase 2, AtAO3, Arabidopsis thaliana aldehyde oxidase 3 |
-0.185 |
0.105 |
| 97 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
-0.184 |
0.103 |
| 98 |
267144_at |
AT2G38110
|
ATGPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6, GPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6 |
-0.182 |
-0.098 |
| 99 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.181 |
-0.171 |