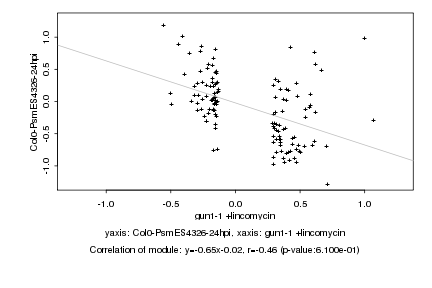
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
gun1-1 +lincomycin |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
256603_at |
AT3G28270
|
unknown |
1.061 |
-0.293 |
| 2 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.992 |
0.992 |
| 3 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
0.709 |
-1.274 |
| 4 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
0.701 |
-0.686 |
| 5 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.662 |
0.489 |
| 6 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.617 |
-0.162 |
| 7 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
0.614 |
0.586 |
| 8 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
0.610 |
-0.609 |
| 9 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.610 |
0.768 |
| 10 |
258025_at |
AT3G19480
|
3-PGDH, PGDH3, phosphoglycerate dehydrogenase 3 |
0.591 |
-0.676 |
| 11 |
264648_at |
AT1G09080
|
BIP3, binding protein 3 |
0.576 |
0.117 |
| 12 |
248890_at |
AT5G46270
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.573 |
-0.054 |
| 13 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
0.572 |
-0.092 |
| 14 |
263597_at |
AT2G01870
|
unknown |
0.536 |
-0.110 |
| 15 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
0.536 |
-0.244 |
| 16 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
0.532 |
-0.685 |
| 17 |
246004_at |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
0.501 |
-0.784 |
| 18 |
251155_at |
AT3G63160
|
OEP6, outer envelope protein 6 |
0.493 |
-0.762 |
| 19 |
252366_at |
AT3G48420
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.486 |
-0.676 |
| 20 |
261430_at |
AT1G18830
|
SEC31A |
0.474 |
0.082 |
| 21 |
259352_at |
AT3G05170
|
[Phosphoglycerate mutase family protein] |
0.471 |
0.288 |
| 22 |
263287_at |
AT2G36145
|
unknown |
0.468 |
-0.733 |
| 23 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
0.467 |
-0.933 |
| 24 |
256015_at |
AT1G19150
|
LHCA2*1, Lhca6, photosystem I light harvesting complex gene 6 |
0.456 |
-0.871 |
| 25 |
249120_at |
AT5G43750
|
NDH18, NAD(P)H dehydrogenase 18, PnsB5, Photosynthetic NDH subcomplex B 5 |
0.453 |
-0.544 |
| 26 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.439 |
-0.667 |
| 27 |
264096_at |
AT1G78995
|
unknown |
0.434 |
-0.560 |
| 28 |
245876_at |
AT1G26230
|
Cpn60beta4, chaperonin-60beta4 |
0.424 |
-0.765 |
| 29 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.420 |
0.840 |
| 30 |
252853_at |
AT4G39710
|
FKBP16-2, FK506-binding protein 16-2, PnsL4, Photosynthetic NDH subcomplex L 4 |
0.411 |
-0.911 |
| 31 |
250506_at |
AT5G09930
|
ABCF2, ATP-binding cassette F2 |
0.410 |
0.174 |
| 32 |
266979_at |
AT2G39470
|
PnsL1, Photosynthetic NDH subcomplex L 1, PPL2, PsbP-like protein 2 |
0.403 |
-0.780 |
| 33 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
0.394 |
0.022 |
| 34 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.393 |
0.190 |
| 35 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
0.390 |
-0.804 |
| 36 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.384 |
-0.411 |
| 37 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
0.378 |
-0.933 |
| 38 |
264435_at |
AT1G10360
|
ATGSTU18, glutathione S-transferase TAU 18, GST29, GLUTATHIONE S-TRANSFERASE 29, GSTU18, glutathione S-transferase TAU 18 |
0.370 |
-0.427 |
| 39 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
0.368 |
-0.883 |
| 40 |
262313_at |
AT1G70900
|
unknown |
0.367 |
0.046 |
| 41 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.358 |
-0.140 |
| 42 |
262288_at |
AT1G70760
|
CRR23, CHLORORESPIRATORY REDUCTION 23, NdhL, NADH dehydrogenase-like complex L |
0.355 |
-0.770 |
| 43 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.347 |
-0.632 |
| 44 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
0.346 |
-0.575 |
| 45 |
258087_at |
AT3G26060
|
ATPRX Q, PRXQ, peroxiredoxin Q |
0.346 |
-0.674 |
| 46 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
0.342 |
0.190 |
| 47 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.335 |
-0.537 |
| 48 |
249510_at |
AT5G38510
|
[Rhomboid-related intramembrane serine protease family protein] |
0.334 |
-0.367 |
| 49 |
261801_at |
AT1G30520
|
AAE14, acyl-activating enzyme 14 |
0.334 |
-0.576 |
| 50 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.328 |
0.320 |
| 51 |
258333_at |
AT3G16000
|
MFP1, MAR binding filament-like protein 1 |
0.327 |
-0.460 |
| 52 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
0.316 |
-0.587 |
| 53 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
0.316 |
-0.450 |
| 54 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
0.311 |
-0.350 |
| 55 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
0.310 |
-0.786 |
| 56 |
250211_at |
AT5G13880
|
unknown |
0.305 |
0.346 |
| 57 |
260309_at |
AT1G70580
|
AOAT2, alanine-2-oxoglutarate aminotransferase 2, GGT2, GLUTAMATE:GLYOXYLATE AMINOTRANSFERASE 2 |
0.305 |
-0.156 |
| 58 |
250802_at |
AT5G04970
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.304 |
0.071 |
| 59 |
267505_at |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
0.300 |
-0.410 |
| 60 |
254398_at |
AT4G21280
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-1, PHOTOSYSTEM II SUBUNIT Q-1, PSBQA, photosystem II subunit QA |
0.295 |
-0.328 |
| 61 |
267483_at |
AT2G02810
|
ATUTR1, UDP-GALACTOSE TRANSPORTER 1, UTR1, UDP-galactose transporter 1 |
0.293 |
0.252 |
| 62 |
248094_at |
AT5G55220
|
[trigger factor type chaperone family protein] |
0.292 |
-0.963 |
| 63 |
247102_at |
AT5G66550
|
[Maf-like protein] |
0.291 |
-0.200 |
| 64 |
247943_at |
AT5G57170
|
[Tautomerase/MIF superfamily protein] |
0.291 |
-0.864 |
| 65 |
256979_at |
AT3G21055
|
PSBTN, photosystem II subunit T |
0.290 |
-0.376 |
| 66 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
0.289 |
-0.634 |
| 67 |
250262_at |
AT5G13410
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.288 |
-0.189 |
| 68 |
250739_at |
AT5G05740
|
ATEGY2, EGY2, ethylene-dependent gravitropism-deficient and yellow-green-like 2 |
0.287 |
-0.529 |
| 69 |
259981_at |
AT1G76450
|
[Photosystem II reaction center PsbP family protein] |
0.286 |
-0.336 |
| 70 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
-0.560 |
1.192 |
| 71 |
260012_at |
AT1G67865
|
unknown |
-0.504 |
0.139 |
| 72 |
266712_at |
AT2G46750
|
AtGulLO2, GulLO2, L -gulono-1,4-lactone ( L -GulL) oxidase 2 |
-0.502 |
-0.034 |
| 73 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.445 |
0.901 |
| 74 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
-0.412 |
1.021 |
| 75 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
-0.395 |
0.433 |
| 76 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
-0.363 |
0.751 |
| 77 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
-0.344 |
0.002 |
| 78 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
-0.319 |
0.237 |
| 79 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
-0.318 |
0.094 |
| 80 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
-0.301 |
-0.030 |
| 81 |
247197_at |
AT5G65240
|
[Leucine-rich repeat protein kinase family protein] |
-0.301 |
0.284 |
| 82 |
246933_at |
AT5G25160
|
ZFP3, zinc finger protein 3 |
-0.294 |
-0.138 |
| 83 |
260004_at |
AT1G67860
|
unknown |
-0.286 |
0.105 |
| 84 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
-0.277 |
0.792 |
| 85 |
245041_at |
AT2G26530
|
AR781 |
-0.272 |
0.467 |
| 86 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
-0.264 |
0.868 |
| 87 |
247425_at |
AT5G62550
|
unknown |
-0.264 |
-0.122 |
| 88 |
246759_at |
AT5G27950
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.255 |
0.038 |
| 89 |
257670_at |
AT3G20340
|
unknown |
-0.255 |
0.300 |
| 90 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
-0.246 |
-0.231 |
| 91 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
-0.231 |
-0.309 |
| 92 |
250872_at |
AT5G03960
|
IQD12, IQ-domain 12 |
-0.231 |
0.252 |
| 93 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
-0.228 |
-0.301 |
| 94 |
249032_at |
AT5G44910
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.224 |
0.093 |
| 95 |
245932_at |
AT5G09290
|
[Inositol monophosphatase family protein] |
-0.222 |
0.525 |
| 96 |
244982_at |
ATCG00780
|
RPL14, ribosomal protein L14 |
-0.215 |
-0.173 |
| 97 |
258787_at |
AT3G11840
|
PUB24, plant U-box 24 |
-0.212 |
0.581 |
| 98 |
255626_at |
AT4G00780
|
[TRAF-like family protein] |
-0.201 |
-0.111 |
| 99 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
-0.199 |
0.239 |
| 100 |
254551_at |
AT4G19840
|
ATPP2-A1, phloem protein 2-A1, ATPP2A-1, phloem protein 2-A1, PP2-A1, phloem protein 2-A1 |
-0.187 |
0.025 |
| 101 |
262782_at |
AT1G13195
|
[RING/U-box superfamily protein] |
-0.183 |
0.310 |
| 102 |
265345_at |
AT2G22680
|
WAVH1, WAV3 homolog 1 |
-0.183 |
0.359 |
| 103 |
261441_at |
AT1G28470
|
ANAC010, NAC domain containing protein 10, NAC010, NAC domain containing protein 10, SND3, SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 3 |
-0.180 |
0.017 |
| 104 |
254347_at |
AT4G22070
|
ATWRKY31, WRKY31, WRKY DNA-binding protein 31 |
-0.180 |
0.572 |
| 105 |
257113_at |
AT3G20130
|
CYP705A22, cytochrome P450, family 705, subfamily A, polypeptide 22, GPS1, gravity persistence signal 1 |
-0.178 |
0.037 |
| 106 |
244981_at |
ATCG00770
|
RPS8, ribosomal protein S8 |
-0.175 |
-0.131 |
| 107 |
262071_at |
AT1G59510
|
CF9 |
-0.172 |
0.050 |
| 108 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.172 |
-0.760 |
| 109 |
254592_at |
AT4G18880
|
AT-HSFA4A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A4A, HSF A4A, heat shock transcription factor A4A |
-0.171 |
0.246 |
| 110 |
263427_at |
AT2G22260
|
ALKBH2, homolog of E. coli alkB |
-0.171 |
-0.109 |
| 111 |
246133_at |
AT5G20960
|
AAO1, aldehyde oxidase 1, AO1, aldehyde oxidase 1, AOalpha, aldehyde oxidase alpha, AT-AO1, ARABIDOPSIS THALIANA ALDEHYDE OXIDASE 1, ATAO, AtAO1, Arabidopsis thaliana aldehyde oxidase 1 |
-0.170 |
0.678 |
| 112 |
247902_at |
AT5G57350
|
AHA3, H(+)-ATPase 3, ATAHA3, ARABIDOPSIS THALIANA ARABIDOPSIS H(+)-ATPASE, HA3, H(+)-ATPase 3 |
-0.169 |
0.065 |
| 113 |
259169_at |
AT3G03520
|
NPC3, non-specific phospholipase C3 |
-0.168 |
0.126 |
| 114 |
245017_at |
ATCG00510
|
PSAI, photsystem I subunit I |
-0.166 |
-0.138 |
| 115 |
248469_at |
AT5G50820
|
anac097, NAC domain containing protein 97, NAC097, NAC domain containing protein 97 |
-0.164 |
-0.036 |
| 116 |
253510_at |
AT4G31730
|
GDU1, glutamine dumper 1 |
-0.163 |
-0.016 |
| 117 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
-0.162 |
0.817 |
| 118 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
-0.160 |
0.017 |
| 119 |
251968_at |
AT3G53100
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.160 |
-0.349 |
| 120 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
-0.160 |
0.464 |
| 121 |
244986_at |
ATCG00820
|
RPS19, ribosomal protein S19 |
-0.159 |
-0.195 |
| 122 |
244965_at |
ATCG00590
|
ORF31 |
-0.157 |
0.020 |
| 123 |
258445_at |
AT3G22400
|
ATLOX5, Arabidopsis thaliana lipoxygenase 5, LOX5 |
-0.157 |
0.274 |
| 124 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
-0.156 |
0.067 |
| 125 |
253493_at |
AT4G31820
|
ENP, ENHANCER OF PINOID, MAB4, MACCHI-BOU 4, NPY1, NAKED PINS IN YUC MUTANTS 1 |
-0.155 |
-0.407 |
| 126 |
258228_at |
AT3G27610
|
[Nucleotidylyl transferase superfamily protein] |
-0.154 |
0.450 |
| 127 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.153 |
-0.228 |
| 128 |
247324_at |
AT5G64190
|
unknown |
-0.153 |
0.471 |
| 129 |
257476_at |
AT1G80960
|
[F-box and Leucine Rich Repeat domains containing protein] |
-0.152 |
0.284 |
| 130 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
-0.152 |
-0.034 |
| 131 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.148 |
-0.005 |
| 132 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
-0.146 |
0.301 |
| 133 |
253036_at |
AT4G38340
|
NLP3, NIN-like protein 3 |
-0.141 |
0.014 |
| 134 |
266375_at |
AT2G14630
|
unknown |
-0.140 |
0.148 |
| 135 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
-0.140 |
-0.731 |
| 136 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.136 |
0.200 |
| 137 |
265440_at |
AT2G20960
|
pEARLI4 |
-0.135 |
0.171 |