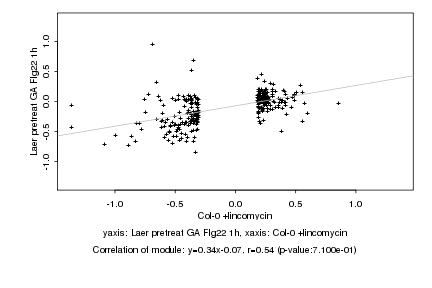
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-0 +lincomycin |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.854 |
-0.031 |
| 2 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
0.595 |
-0.189 |
| 3 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.572 |
-0.026 |
| 4 |
260668_at |
AT1G19530
|
unknown |
0.551 |
0.158 |
| 5 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.549 |
-0.329 |
| 6 |
261430_at |
AT1G18830
|
SEC31A |
0.538 |
0.268 |
| 7 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.502 |
0.167 |
| 8 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.488 |
0.132 |
| 9 |
256188_at |
AT1G30160
|
unknown |
0.487 |
0.121 |
| 10 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
0.484 |
0.032 |
| 11 |
247317_at |
AT5G64060
|
anac103, NAC domain containing protein 103, NAC103, NAC domain containing protein 103 |
0.484 |
0.105 |
| 12 |
263515_at |
AT2G21640
|
unknown |
0.473 |
0.077 |
| 13 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.462 |
-0.095 |
| 14 |
263118_at |
AT1G03090
|
MCCA |
0.430 |
0.051 |
| 15 |
244997_at |
ATCG00170
|
RPOC2 |
0.419 |
-0.207 |
| 16 |
267279_at |
AT2G19460
|
unknown |
0.415 |
-0.059 |
| 17 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
0.413 |
-0.014 |
| 18 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
0.408 |
0.126 |
| 19 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.405 |
0.174 |
| 20 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
0.402 |
-0.012 |
| 21 |
259852_at |
AT1G72280
|
AERO1, endoplasmic reticulum oxidoreductins 1, ERO1, endoplasmic reticulum oxidoreductins 1 |
0.391 |
0.186 |
| 22 |
259388_at |
AT1G13420
|
ATST4B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4B, ST4B, sulfotransferase 4B |
0.389 |
0.018 |
| 23 |
251022_at |
AT5G02150
|
Fes1C, Fes1C |
0.386 |
-0.111 |
| 24 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
0.381 |
-0.498 |
| 25 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
0.375 |
-0.002 |
| 26 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.374 |
0.038 |
| 27 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.362 |
-0.000 |
| 28 |
244982_at |
ATCG00780
|
RPL14, ribosomal protein L14 |
0.356 |
-0.064 |
| 29 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
0.354 |
-0.099 |
| 30 |
249870_at |
AT5G23080
|
TGH, TOUGH |
0.351 |
0.053 |
| 31 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.332 |
0.179 |
| 32 |
244986_at |
ATCG00820
|
RPS19, ribosomal protein S19 |
0.327 |
-0.090 |
| 33 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.322 |
-0.016 |
| 34 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
0.314 |
0.008 |
| 35 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.314 |
0.299 |
| 36 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.305 |
0.085 |
| 37 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.303 |
0.085 |
| 38 |
261167_at |
AT1G04980
|
ATPDI10, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 10, ATPDIL2-2, PDI-like 2-2, PDI10, PROTEIN DISULFIDE ISOMERASE, PDIL2-2, PDI-like 2-2 |
0.303 |
0.217 |
| 39 |
254174_at |
AT4G24120
|
ATYSL1, YELLOW STRIPE LIKE 1, YSL1, YELLOW STRIPE like 1 |
0.303 |
-0.131 |
| 40 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.302 |
0.126 |
| 41 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
0.301 |
0.179 |
| 42 |
244998_at |
ATCG00180
|
RPOC1 |
0.284 |
-0.125 |
| 43 |
262888_at |
AT1G14790
|
AtRDR1, ATRDRP1, RDR1, RNA-dependent RNA polymerase 1 |
0.284 |
-0.053 |
| 44 |
261947_at |
AT1G64470
|
[Ubiquitin-like superfamily protein] |
0.283 |
0.315 |
| 45 |
259982_at |
AT1G76410
|
ATL8 |
0.277 |
0.046 |
| 46 |
247425_at |
AT5G62550
|
unknown |
0.271 |
0.027 |
| 47 |
252539_at |
AT3G45730
|
unknown |
0.270 |
-0.028 |
| 48 |
244929_at |
ATMG00580
|
NAD4, NADH dehydrogenase subunit 4 |
0.270 |
-0.003 |
| 49 |
246906_at |
AT5G25475
|
[AP2/B3-like transcriptional factor family protein] |
0.270 |
0.026 |
| 50 |
259173_at |
AT3G03640
|
BGLU25, beta glucosidase 25, GLUC |
0.269 |
0.070 |
| 51 |
255703_at |
AT4G00040
|
[Chalcone and stilbene synthase family protein] |
0.266 |
0.078 |
| 52 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.265 |
-0.159 |
| 53 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.264 |
0.085 |
| 54 |
262124_at |
AT1G59660
|
[Nucleoporin autopeptidase] |
0.262 |
0.158 |
| 55 |
251373_at |
AT3G60530
|
GATA4, GATA transcription factor 4 |
0.261 |
-0.116 |
| 56 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.256 |
0.209 |
| 57 |
263707_at |
AT1G09300
|
[Metallopeptidase M24 family protein] |
0.255 |
0.022 |
| 58 |
250572_at |
AT5G08210
|
MIR834A, microRNA834A |
0.253 |
0.024 |
| 59 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
0.251 |
0.050 |
| 60 |
253279_at |
AT4G34030
|
MCCB, 3-methylcrotonyl-CoA carboxylase |
0.250 |
0.016 |
| 61 |
256425_at |
AT1G33560
|
ADR1, ACTIVATED DISEASE RESISTANCE 1 |
0.250 |
-0.055 |
| 62 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.246 |
0.140 |
| 63 |
248302_at |
AT5G53160
|
PYL8, PYR1-like 8, RCAR3, regulatory components of ABA receptor 3 |
0.246 |
-0.030 |
| 64 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
0.246 |
0.129 |
| 65 |
251035_at |
AT5G02220
|
SMR4, SIAMESE-RELATED 4 |
0.246 |
0.209 |
| 66 |
262255_at |
AT1G53790
|
[F-box and associated interaction domains-containing protein] |
0.244 |
0.089 |
| 67 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.241 |
0.064 |
| 68 |
249058_at |
AT5G44510
|
TAO1, target of AVRB operation1 |
0.239 |
0.109 |
| 69 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.239 |
0.108 |
| 70 |
260687_at |
AT1G17530
|
ATTIM23-1, translocase of inner mitochondrial membrane 23, TIM23-1, translocase of inner mitochondrial membrane 23 |
0.236 |
0.081 |
| 71 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
0.235 |
0.335 |
| 72 |
248204_at |
AT5G54280
|
ATM2, myosin 2, ATM4, ARABIDOPSIS THALIANA MYOSIN 4, ATMYOS1, ARABIDOPSIS THALIANA MYOSIN 1 |
0.234 |
0.085 |
| 73 |
263839_at |
AT2G36900
|
ATMEMB11, MEMB11, membrin 11 |
0.234 |
0.168 |
| 74 |
248134_at |
AT5G54860
|
[Major facilitator superfamily protein] |
0.233 |
0.124 |
| 75 |
266428_at |
AT2G07180
|
[Protein kinase superfamily protein] |
0.233 |
0.143 |
| 76 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
0.233 |
-0.002 |
| 77 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.233 |
-0.062 |
| 78 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.231 |
-0.021 |
| 79 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
0.228 |
-0.115 |
| 80 |
260740_at |
AT1G15020
|
ATQSOX1, quiescin-sulfhydryl oxidase 1, QSO2, QSOX1, quiescin-sulfhydryl oxidase 1 |
0.228 |
0.155 |
| 81 |
262845_at |
AT1G14740
|
TTA1, TITANIA 1 |
0.226 |
0.062 |
| 82 |
251864_at |
AT3G54920
|
PMR6, powdery mildew resistant 6 |
0.226 |
-0.308 |
| 83 |
264752_at |
AT1G23010
|
LPR1, Low Phosphate Root1 |
0.226 |
0.031 |
| 84 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
0.224 |
0.070 |
| 85 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
0.224 |
-0.163 |
| 86 |
258331_at |
AT3G15980
|
[Coatomer, beta' subunit] |
0.223 |
0.179 |
| 87 |
263633_at |
AT2G04890
|
SCL21, SCARECROW-like 21 |
0.223 |
-0.109 |
| 88 |
265383_at |
AT2G16780
|
MSI02, MSI2, MULTICOPY SUPPRESSOR OF IRA1 2, NFC02, NFC2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 2 |
0.221 |
0.054 |
| 89 |
251641_at |
AT3G57470
|
[Insulinase (Peptidase family M16) family protein] |
0.219 |
-0.145 |
| 90 |
244996_at |
ATCG00160
|
RPS2, ribosomal protein S2 |
0.219 |
-0.122 |
| 91 |
251138_at |
AT5G01160
|
[RING/U-box superfamily protein] |
0.218 |
0.069 |
| 92 |
248228_at |
AT5G53800
|
unknown |
0.217 |
0.138 |
| 93 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
0.216 |
-0.012 |
| 94 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.216 |
0.083 |
| 95 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
0.215 |
0.203 |
| 96 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.215 |
0.061 |
| 97 |
255472_at |
AT4G02430
|
At-SR34b, Serine/Arginine-Rich Protein Splicing Factor 34b, SR34b, Serine/Arginine-Rich Protein Splicing Factor 34b |
0.213 |
0.195 |
| 98 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
0.209 |
0.459 |
| 99 |
244937_at |
ATCG01110
|
NDHH, NAD(P)H dehydrogenase subunit H |
0.209 |
-0.095 |
| 100 |
259061_at |
AT3G07410
|
AtRABA5b, RAB GTPase homolog A5B, RABA5b, RAB GTPase homolog A5B |
0.208 |
0.014 |
| 101 |
253074_at |
AT4G36140
|
[disease resistance protein (TIR-NBS-LRR class), putative] |
0.207 |
0.076 |
| 102 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
0.206 |
0.105 |
| 103 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
0.205 |
-0.350 |
| 104 |
246506_at |
AT5G16110
|
unknown |
0.205 |
0.058 |
| 105 |
253175_at |
AT4G35050
|
MSI3, MULTICOPY SUPPRESSOR OF IRA1 3, NFC3, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 3 |
0.205 |
0.002 |
| 106 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
0.204 |
0.143 |
| 107 |
250911_at |
AT5G03730
|
AtCTR1, CTR1, CONSTITUTIVE TRIPLE RESPONSE 1, SIS1, SUGAR-INSENSITIVE 1 |
0.202 |
0.052 |
| 108 |
265481_at |
AT2G15960
|
unknown |
0.202 |
0.072 |
| 109 |
257940_at |
AT3G21790
|
[UDP-Glycosyltransferase superfamily protein] |
0.202 |
0.076 |
| 110 |
253388_at |
AT4G32910
|
unknown |
0.199 |
-0.110 |
| 111 |
252095_at |
AT3G51000
|
[alpha/beta-Hydrolases superfamily protein] |
0.199 |
0.055 |
| 112 |
254703_at |
AT4G17960
|
unknown |
0.198 |
0.038 |
| 113 |
255526_at |
AT4G02350
|
SEC15B, exocyst complex component sec15B |
0.198 |
0.176 |
| 114 |
253958_at |
AT4G26400
|
[RING/U-box superfamily protein] |
0.195 |
0.209 |
| 115 |
245117_at |
AT2G41560
|
ACA4, autoinhibited Ca(2+)-ATPase, isoform 4 |
0.195 |
-0.323 |
| 116 |
259230_at |
AT3G07780
|
OBE1, OBERON1 |
0.194 |
0.073 |
| 117 |
247284_at |
AT5G64410
|
ATOPT4, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 4, OPT4, oligopeptide transporter 4 |
0.193 |
-0.030 |
| 118 |
266511_at |
AT2G47680
|
[zinc finger (CCCH type) helicase family protein] |
0.193 |
0.069 |
| 119 |
262395_at |
AT1G49540
|
AtELP2, Elongator subunit 2, ELP2, elongator protein 2 |
0.193 |
0.043 |
| 120 |
258114_at |
AT3G14660
|
CYP72A13, cytochrome P450, family 72, subfamily A, polypeptide 13 |
0.191 |
-0.169 |
| 121 |
265337_at |
AT2G18390
|
ARL2, ARF-LIKE 2, ATARLC1, HAL, HALLIMASCH, TTN5, TITAN 5 |
0.190 |
0.083 |
| 122 |
253702_at |
AT4G29900
|
ACA10, autoinhibited Ca(2+)-ATPase 10, ATACA10, CIF1, COMPACT INFLORESCENCE 1 |
0.190 |
0.074 |
| 123 |
251090_at |
AT5G01340
|
AtmSFC1, mSFC1, mitochondrial succinate-fumarate carrier 1 |
0.189 |
0.109 |
| 124 |
251554_at |
AT3G58670
|
unknown |
0.189 |
-0.079 |
| 125 |
260112_at |
AT1G63310
|
unknown |
0.189 |
-0.074 |
| 126 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.189 |
-0.261 |
| 127 |
264658_at |
AT1G09910
|
[Rhamnogalacturonate lyase family protein] |
0.189 |
-0.045 |
| 128 |
250737_at |
AT5G06370
|
[NC domain-containing protein-related] |
0.188 |
0.014 |
| 129 |
247043_at |
AT5G66880
|
SNRK2-3, SUCROSE NONFERMENTING 1 (SNF1)-RELATED PROTEIN KINASE 2-3, SNRK2.3, sucrose nonfermenting 1(SNF1)-related protein kinase 2.3, SRK2I |
0.187 |
0.013 |
| 130 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
0.186 |
0.125 |
| 131 |
260502_at |
AT1G47270
|
AtTLP6, tubby like protein 6, TLP6, tubby like protein 6 |
0.186 |
0.173 |
| 132 |
247748_at |
AT5G58920
|
unknown |
0.186 |
0.051 |
| 133 |
250419_at |
AT5G11250
|
[Disease resistance protein (TIR-NBS-LRR class)] |
0.185 |
-0.161 |
| 134 |
258630_at |
AT3G02820
|
[zinc knuckle (CCHC-type) family protein] |
0.185 |
0.214 |
| 135 |
246184_at |
AT5G20950
|
[Glycosyl hydrolase family protein] |
0.185 |
-0.092 |
| 136 |
246055_at |
AT5G08380
|
AGAL1, alpha-galactosidase 1, AtAGAL1, alpha-galactosidase 1 |
0.184 |
-0.191 |
| 137 |
261654_at |
AT1G01920
|
[SET domain-containing protein] |
0.184 |
0.055 |
| 138 |
255221_at |
AT4G05150
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.183 |
0.110 |
| 139 |
261521_at |
AT1G71830
|
ATSERK1, SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE 1, SERK1, somatic embryogenesis receptor-like kinase 1 |
0.182 |
0.392 |
| 140 |
245359_at |
AT4G14430
|
ATECI2, ARABIDOPSIS THALIANA DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 2, ECHIB, ENOYL-COA HYDRATASE/ISOMERASE B, ECI2, DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 2, IBR10, indole-3-butyric acid response 10, PEC12 |
0.182 |
0.079 |
| 141 |
262770_at |
AT1G13190
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.181 |
0.025 |
| 142 |
245261_at |
AT4G14385
|
unknown |
0.180 |
-0.014 |
| 143 |
256603_at |
AT3G28270
|
unknown |
-1.367 |
-0.432 |
| 144 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-1.364 |
-0.050 |
| 145 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-1.089 |
-0.714 |
| 146 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-1.003 |
-0.560 |
| 147 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.888 |
-0.724 |
| 148 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.866 |
-0.575 |
| 149 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.830 |
-0.654 |
| 150 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.826 |
-0.362 |
| 151 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.799 |
-0.355 |
| 152 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.781 |
-0.461 |
| 153 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.763 |
0.045 |
| 154 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.751 |
-0.181 |
| 155 |
249476_at |
AT5G38910
|
[RmlC-like cupins superfamily protein] |
-0.722 |
0.123 |
| 156 |
251755_at |
AT3G55790
|
unknown |
-0.696 |
0.960 |
| 157 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
-0.662 |
0.326 |
| 158 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.657 |
-0.292 |
| 159 |
246004_at |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
-0.640 |
0.086 |
| 160 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
-0.625 |
0.032 |
| 161 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.617 |
-0.420 |
| 162 |
258025_at |
AT3G19480
|
3-PGDH, PGDH3, phosphoglycerate dehydrogenase 3 |
-0.614 |
-0.331 |
| 163 |
251155_at |
AT3G63160
|
OEP6, outer envelope protein 6 |
-0.611 |
-0.300 |
| 164 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.608 |
-0.192 |
| 165 |
263715_at |
AT2G20570
|
GBF's pro-rich region-interacting factor 1 |
-0.602 |
-0.060 |
| 166 |
265892_at |
AT2G15020
|
unknown |
-0.596 |
-0.402 |
| 167 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.594 |
-0.597 |
| 168 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.581 |
-0.343 |
| 169 |
252853_at |
AT4G39710
|
FKBP16-2, FK506-binding protein 16-2, PnsL4, Photosynthetic NDH subcomplex L 4 |
-0.578 |
-0.545 |
| 170 |
263597_at |
AT2G01870
|
unknown |
-0.565 |
-0.298 |
| 171 |
256096_at |
AT1G13650
|
unknown |
-0.563 |
-0.647 |
| 172 |
256015_at |
AT1G19150
|
LHCA2*1, Lhca6, photosystem I light harvesting complex gene 6 |
-0.555 |
-0.502 |
| 173 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.555 |
-0.498 |
| 174 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.542 |
-0.403 |
| 175 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.540 |
-0.397 |
| 176 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-0.529 |
-0.349 |
| 177 |
263674_at |
AT2G04790
|
unknown |
-0.529 |
-0.694 |
| 178 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.523 |
0.054 |
| 179 |
250613_at |
AT5G07240
|
IQD24, IQ-domain 24 |
-0.522 |
-0.568 |
| 180 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
-0.514 |
-0.244 |
| 181 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.511 |
-0.390 |
| 182 |
264839_at |
AT1G03630
|
POR C, protochlorophyllide oxidoreductase C, PORC |
-0.510 |
-0.386 |
| 183 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
-0.499 |
0.030 |
| 184 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.498 |
-0.374 |
| 185 |
249120_at |
AT5G43750
|
NDH18, NAD(P)H dehydrogenase 18, PnsB5, Photosynthetic NDH subcomplex B 5 |
-0.497 |
-0.578 |
| 186 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.490 |
-0.488 |
| 187 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.487 |
-0.451 |
| 188 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.478 |
0.040 |
| 189 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.477 |
0.106 |
| 190 |
247816_at |
AT5G58260
|
NdhN, NADH dehydrogenase-like complex N |
-0.476 |
-0.393 |
| 191 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.476 |
-0.428 |
| 192 |
262175_at |
AT1G74880
|
NDH-O, NAD(P)H:plastoquinone dehydrogenase complex subunit O, NdhO, NADH dehydrogenase-like complex ) |
-0.470 |
-0.170 |
| 193 |
252366_at |
AT3G48420
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.470 |
-0.643 |
| 194 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
-0.470 |
-0.466 |
| 195 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.459 |
-0.270 |
| 196 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.458 |
-0.379 |
| 197 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
-0.451 |
-0.522 |
| 198 |
266979_at |
AT2G39470
|
PnsL1, Photosynthetic NDH subcomplex L 1, PPL2, PsbP-like protein 2 |
-0.448 |
-0.600 |
| 199 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
-0.447 |
-0.347 |
| 200 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
-0.434 |
0.084 |
| 201 |
261118_at |
AT1G75460
|
[ATP-dependent protease La (LON) domain protein] |
-0.430 |
-0.074 |
| 202 |
261075_at |
AT1G07280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.430 |
0.044 |
| 203 |
252441_at |
AT3G46780
|
PTAC16, plastid transcriptionally active 16 |
-0.428 |
-0.367 |
| 204 |
245209_at |
AT5G12340
|
unknown |
-0.417 |
0.026 |
| 205 |
256655_at |
AT3G18890
|
AtTic62, translocon at the inner envelope membrane of chloroplasts 62, Tic62, translocon at the inner envelope membrane of chloroplasts 62 |
-0.415 |
-0.344 |
| 206 |
245876_at |
AT1G26230
|
Cpn60beta4, chaperonin-60beta4 |
-0.415 |
-0.544 |
| 207 |
262313_at |
AT1G70900
|
unknown |
-0.414 |
-0.339 |
| 208 |
267359_at |
AT2G40020
|
[Nucleolar histone methyltransferase-related protein] |
-0.409 |
0.003 |
| 209 |
262288_at |
AT1G70760
|
CRR23, CHLORORESPIRATORY REDUCTION 23, NdhL, NADH dehydrogenase-like complex L |
-0.408 |
-0.660 |
| 210 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
-0.407 |
0.047 |
| 211 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.398 |
-0.584 |
| 212 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.398 |
-0.188 |
| 213 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.397 |
-0.397 |
| 214 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
-0.397 |
0.042 |
| 215 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.396 |
0.113 |
| 216 |
255719_at |
AT1G32080
|
AtLrgB, LrgB |
-0.395 |
-0.267 |
| 217 |
258333_at |
AT3G16000
|
MFP1, MAR binding filament-like protein 1 |
-0.394 |
-0.142 |
| 218 |
255046_at |
AT4G09650
|
ATPD, ATP synthase delta-subunit gene, PDE332, PIGMENT DEFECTIVE 332 |
-0.394 |
-0.178 |
| 219 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.381 |
-0.354 |
| 220 |
259707_at |
AT1G77490
|
TAPX, thylakoidal ascorbate peroxidase |
-0.381 |
-0.323 |
| 221 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
-0.380 |
-0.028 |
| 222 |
266278_at |
AT2G29300
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.377 |
-0.196 |
| 223 |
259188_at |
AT3G01510
|
LSF1, like SEX4 1 |
-0.376 |
0.084 |
| 224 |
267057_at |
AT2G32500
|
[Stress responsive alpha-beta barrel domain protein] |
-0.374 |
-0.327 |
| 225 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
-0.373 |
0.526 |
| 226 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
-0.372 |
-0.305 |
| 227 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
-0.372 |
-0.212 |
| 228 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.372 |
-0.494 |
| 229 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.372 |
0.045 |
| 230 |
249524_at |
AT5G38520
|
[alpha/beta-Hydrolases superfamily protein] |
-0.371 |
-0.246 |
| 231 |
254398_at |
AT4G21280
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-1, PHOTOSYSTEM II SUBUNIT Q-1, PSBQA, photosystem II subunit QA |
-0.371 |
-0.202 |
| 232 |
258087_at |
AT3G26060
|
ATPRX Q, PRXQ, peroxiredoxin Q |
-0.370 |
-0.318 |
| 233 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.367 |
0.023 |
| 234 |
249510_at |
AT5G38510
|
[Rhomboid-related intramembrane serine protease family protein] |
-0.367 |
-0.022 |
| 235 |
247415_at |
AT5G63060
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.367 |
-0.094 |
| 236 |
266655_at |
AT2G25880
|
AtAUR2, ataurora2, AUR2, ataurora2 |
-0.366 |
0.061 |
| 237 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.366 |
0.027 |
| 238 |
245130_at |
AT2G45340
|
[Leucine-rich repeat protein kinase family protein] |
-0.364 |
-0.289 |
| 239 |
253166_at |
AT4G35290
|
ATGLR3.2, ATGLUR2, GLR3.2, GLUTAMATE RECEPTOR 3.2, GLUR2, glutamate receptor 2 |
-0.362 |
-0.028 |
| 240 |
257801_at |
AT3G18750
|
ATWNK6, ARABIDOPSIS THALIANA WITH NO K 6, WNK6, with no lysine (K) kinase 6, ZIK5 |
-0.355 |
-0.309 |
| 241 |
256979_at |
AT3G21055
|
PSBTN, photosystem II subunit T |
-0.355 |
-0.162 |
| 242 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
-0.355 |
0.700 |
| 243 |
245877_at |
AT1G26220
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.354 |
-0.476 |
| 244 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.350 |
-0.356 |
| 245 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.349 |
-0.275 |
| 246 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.349 |
-0.659 |
| 247 |
260309_at |
AT1G70580
|
AOAT2, alanine-2-oxoglutarate aminotransferase 2, GGT2, GLUTAMATE:GLYOXYLATE AMINOTRANSFERASE 2 |
-0.347 |
-0.322 |
| 248 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.346 |
-0.593 |
| 249 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
-0.345 |
-0.227 |
| 250 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.344 |
0.101 |
| 251 |
260542_at |
AT2G43560
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.337 |
-0.175 |
| 252 |
262945_at |
AT1G79510
|
[Uncharacterized conserved protein (DUF2358)] |
-0.336 |
-0.002 |
| 253 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.336 |
0.071 |
| 254 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.335 |
-0.847 |
| 255 |
245793_at |
AT1G32220
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.332 |
-0.188 |
| 256 |
255440_at |
AT4G02530
|
[chloroplast thylakoid lumen protein] |
-0.332 |
-0.303 |
| 257 |
266636_at |
AT2G35370
|
GDCH, glycine decarboxylase complex H |
-0.332 |
-0.227 |
| 258 |
264435_at |
AT1G10360
|
ATGSTU18, glutathione S-transferase TAU 18, GST29, GLUTATHIONE S-TRANSFERASE 29, GSTU18, glutathione S-transferase TAU 18 |
-0.330 |
-0.218 |
| 259 |
254910_at |
AT4G11175
|
[Nucleic acid-binding, OB-fold-like protein] |
-0.329 |
-0.300 |
| 260 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
-0.329 |
-0.282 |
| 261 |
265704_at |
AT2G03420
|
unknown |
-0.329 |
-0.382 |
| 262 |
245479_at |
AT4G16140
|
[proline-rich family protein] |
-0.327 |
-0.033 |
| 263 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.327 |
-0.470 |
| 264 |
263142_at |
AT1G65230
|
[Uncharacterized conserved protein (DUF2358)] |
-0.326 |
-0.319 |
| 265 |
250248_at |
AT5G13740
|
ZIF1, zinc induced facilitator 1 |
-0.323 |
0.063 |
| 266 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.323 |
-0.088 |
| 267 |
259840_at |
AT1G52230
|
PSAH-2, PHOTOSYSTEM I SUBUNIT H-2, PSAH2, photosystem I subunit H2, PSI-H |
-0.322 |
-0.157 |
| 268 |
267635_at |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.322 |
-0.331 |
| 269 |
256518_at |
AT1G66080
|
unknown |
-0.320 |
0.048 |
| 270 |
250095_at |
AT5G17230
|
PSY, PHYTOENE SYNTHASE |
-0.319 |
-0.234 |
| 271 |
252463_at |
AT3G47070
|
[LOCATED IN: thylakoid, chloroplast thylakoid membrane, chloroplast, chloroplast envelope] |
-0.319 |
-0.197 |
| 272 |
258386_at |
AT3G15520
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.318 |
-0.296 |
| 273 |
250985_at |
AT5G02830
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.318 |
-0.341 |
| 274 |
265819_at |
AT2G17972
|
unknown |
-0.317 |
-0.225 |
| 275 |
246313_at |
AT1G31920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.317 |
-0.461 |
| 276 |
261218_at |
AT1G20020
|
ATLFNR2, LEAF FNR 2, FNR2, ferredoxin-NADP(+)-oxidoreductase 2, LFNR2, leaf-type chloroplast-targeted FNR 2 |
-0.317 |
-0.004 |
| 277 |
262168_at |
AT1G74730
|
unknown |
-0.317 |
-0.220 |
| 278 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
-0.314 |
-0.047 |
| 279 |
251476_at |
AT3G59670
|
unknown |
-0.312 |
-0.224 |
| 280 |
264597_at |
AT1G04620
|
HCAR, 7-hydroxymethyl chlorophyll a (HMChl) reductase |
-0.310 |
-0.171 |
| 281 |
262086_at |
AT1G56050
|
[GTP-binding protein-related] |
-0.308 |
-0.265 |
| 282 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
-0.308 |
-0.147 |
| 283 |
259037_at |
AT3G09350
|
Fes1A, Fes1A |
-0.308 |
0.046 |