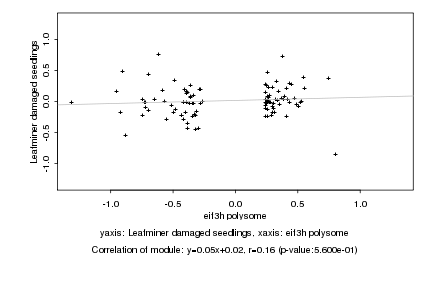
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
eif3h polysome |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
254574_at |
AT4G19430
|
unknown |
0.802 |
-0.848 |
| 2 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.744 |
0.373 |
| 3 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.553 |
0.210 |
| 4 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.545 |
0.387 |
| 5 |
254566_at |
AT4G19240
|
unknown |
0.529 |
0.009 |
| 6 |
250135_at |
AT5G15360
|
unknown |
0.520 |
-0.011 |
| 7 |
247640_at |
AT5G60610
|
[F-box/RNI-like superfamily protein] |
0.501 |
-0.068 |
| 8 |
261930_at |
AT1G22440
|
[Zinc-binding alcohol dehydrogenase family protein] |
0.483 |
-0.037 |
| 9 |
262615_at |
AT1G13950
|
ATELF5A-1, EUKARYOTIC ELONGATION FACTOR 5A-1, EIF-5A, EIF5A, EUKARYOTIC ELONGATION FACTOR 5A, ELF5A-1, eukaryotic elongation factor 5A-1 |
0.470 |
0.064 |
| 10 |
254190_at |
AT4G23885
|
unknown |
0.449 |
0.286 |
| 11 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
0.433 |
-0.004 |
| 12 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
0.433 |
0.291 |
| 13 |
259881_at |
AT1G76820
|
[eukaryotic translation initiation factor 2 (eIF-2) family protein] |
0.417 |
0.034 |
| 14 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.409 |
0.212 |
| 15 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.405 |
-0.227 |
| 16 |
256852_at |
AT3G18610
|
ATNUC-L2, nucleolin like 2, NUC-L2, nucleolin like 2, PARLL1, PARALLEL1-LIKE 1 |
0.386 |
0.087 |
| 17 |
258380_at |
AT3G16650
|
PRL2, Pleiotropic regulatory locus 2 |
0.380 |
0.042 |
| 18 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.370 |
0.726 |
| 19 |
255283_at |
AT4G04620
|
ATG8B, autophagy 8b |
0.364 |
0.063 |
| 20 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
0.351 |
-0.040 |
| 21 |
262003_at |
AT1G64460
|
[Protein kinase superfamily protein] |
0.341 |
0.168 |
| 22 |
261004_at |
AT1G26450
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.335 |
0.027 |
| 23 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.327 |
0.329 |
| 24 |
254854_at |
AT4G12130
|
[Glycine cleavage T-protein family] |
0.314 |
0.037 |
| 25 |
259761_at |
AT1G77590
|
LACS9, long chain acyl-CoA synthetase 9 |
0.311 |
-0.162 |
| 26 |
254350_at |
AT4G22280
|
[F-box/RNI-like superfamily protein] |
0.300 |
-0.104 |
| 27 |
259735_at |
AT1G64405
|
unknown |
0.298 |
-0.028 |
| 28 |
248614_at |
AT5G49560
|
[Putative methyltransferase family protein] |
0.296 |
-0.168 |
| 29 |
256797_at |
AT3G18600
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.294 |
-0.072 |
| 30 |
258707_at |
AT3G09480
|
[Histone superfamily protein] |
0.293 |
0.230 |
| 31 |
257217_at |
AT3G14940
|
ATPPC3, phosphoenolpyruvate carboxylase 3, PPC3, phosphoenolpyruvate carboxylase 3 |
0.286 |
-0.212 |
| 32 |
258623_at |
AT3G02790
|
MBS1, METHYLENE BLUE SENSITIVITY 1 |
0.280 |
-0.007 |
| 33 |
263369_at |
AT2G20480
|
unknown |
0.273 |
-0.012 |
| 34 |
247659_at |
AT5G60040
|
NRPC1, nuclear RNA polymerase C1 |
0.270 |
0.108 |
| 35 |
245147_at |
AT2G45280
|
ATRAD51C, RAS associated with diabetes protein 51C, RAD51C, RAS associated with diabetes protein 51C |
0.263 |
0.067 |
| 36 |
261898_at |
AT1G80720
|
[Mitochondrial glycoprotein family protein] |
0.262 |
0.007 |
| 37 |
251346_at |
AT3G60980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.262 |
0.235 |
| 38 |
248021_at |
AT5G56500
|
Cpn60beta3, chaperonin-60beta3 |
0.254 |
-0.123 |
| 39 |
255535_at |
AT4G01790
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
0.253 |
0.090 |
| 40 |
258079_at |
AT3G25940
|
[TFIIB zinc-binding protein] |
0.252 |
0.013 |
| 41 |
255481_at |
AT4G02460
|
AtPMS1, PMS1, POSTMEIOTIC SEGREGATION 1 |
0.252 |
-0.228 |
| 42 |
247739_at |
AT5G59240
|
[Ribosomal protein S8e family protein] |
0.251 |
0.028 |
| 43 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
0.249 |
0.473 |
| 44 |
246733_at |
AT5G27660
|
DEG14, degradation of periplasmic proteins 14 |
0.247 |
-0.020 |
| 45 |
247412_at |
AT5G63010
|
[Transducin/WD40 repeat-like superfamily protein] |
0.243 |
0.259 |
| 46 |
255224_at |
AT4G05400
|
[copper ion binding] |
0.237 |
-0.236 |
| 47 |
265442_at |
AT2G20940
|
unknown |
0.236 |
-0.064 |
| 48 |
246009_at |
AT5G08335
|
ATICMTB, ARABIDOPSIS THALIANA ISOPRENYL CYSTEINE METHYLTRANSFERASE B, ATSTE14B, ICMTB, ISOPRENYL CYSTEINE METHYLTRANSFERASE B |
0.236 |
0.156 |
| 49 |
247969_at |
AT5G56700
|
[FBD / Leucine Rich Repeat domains containing protein] |
0.235 |
0.038 |
| 50 |
260331_at |
AT1G80270
|
PPR596, PENTATRICOPEPTIDE REPEAT 596 |
0.234 |
-0.108 |
| 51 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.233 |
0.287 |
| 52 |
263712_at |
AT2G20585
|
NFD6, NUCLEAR FUSION DEFECTIVE 6 |
0.233 |
-0.005 |
| 53 |
252462_at |
AT3G47250
|
unknown |
-1.320 |
-0.005 |
| 54 |
260012_at |
AT1G67865
|
unknown |
-0.955 |
0.169 |
| 55 |
265894_at |
AT2G15050
|
LTP, lipid transfer protein, LTP7, lipid transfer protein 7 |
-0.925 |
-0.173 |
| 56 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
-0.912 |
0.490 |
| 57 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
-0.887 |
-0.543 |
| 58 |
254585_at |
AT4G19500
|
[nucleoside-triphosphatases] |
-0.749 |
-0.215 |
| 59 |
263023_at |
AT1G23960
|
unknown |
-0.747 |
0.040 |
| 60 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.732 |
-0.005 |
| 61 |
262790_at |
AT1G10840
|
TIF3H1, translation initiation factor 3 subunit H1 |
-0.723 |
-0.011 |
| 62 |
254251_at |
AT4G23300
|
CRK22, cysteine-rich RLK (RECEPTOR-like protein kinase) 22 |
-0.721 |
-0.090 |
| 63 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.700 |
0.446 |
| 64 |
262458_at |
AT1G11280
|
[S-locus lectin protein kinase family protein] |
-0.699 |
-0.129 |
| 65 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
-0.654 |
0.048 |
| 66 |
259384_at |
AT3G16450
|
JAL33, Jacalin-related lectin 33 |
-0.620 |
0.758 |
| 67 |
248415_at |
AT5G51620
|
[Uncharacterised protein family (UPF0172)] |
-0.589 |
0.178 |
| 68 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.575 |
0.011 |
| 69 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.559 |
-0.282 |
| 70 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.515 |
-0.064 |
| 71 |
245952_at |
AT5G28500
|
unknown |
-0.499 |
-0.163 |
| 72 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
-0.496 |
0.347 |
| 73 |
249124_at |
AT5G43740
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.486 |
-0.117 |
| 74 |
261602_at |
AT1G49630
|
ATPREP2, presequence protease 2, PREP2, presequence protease 2 |
-0.440 |
-0.218 |
| 75 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.433 |
-0.212 |
| 76 |
249242_at |
AT5G42250
|
[Zinc-binding alcohol dehydrogenase family protein] |
-0.420 |
-0.012 |
| 77 |
254870_at |
AT4G11900
|
[S-locus lectin protein kinase family protein] |
-0.419 |
-0.007 |
| 78 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.417 |
-0.279 |
| 79 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
-0.413 |
0.194 |
| 80 |
262563_at |
AT1G34210
|
ATSERK2, SERK2, somatic embryogenesis receptor-like kinase 2 |
-0.402 |
-0.167 |
| 81 |
262750_at |
AT1G28710
|
[Nucleotide-diphospho-sugar transferase family protein] |
-0.399 |
-0.008 |
| 82 |
249486_at |
AT5G39030
|
[Protein kinase superfamily protein] |
-0.399 |
0.176 |
| 83 |
259685_at |
AT1G63090
|
AtPP2-A11, phloem protein 2-A11, PP2-A11, phloem protein 2-A11 |
-0.396 |
0.142 |
| 84 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.392 |
0.150 |
| 85 |
254638_at |
AT4G18740
|
[Rho termination factor] |
-0.386 |
-0.339 |
| 86 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.386 |
-0.428 |
| 87 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.375 |
-0.017 |
| 88 |
247345_at |
AT5G63760
|
ARI15, ARIADNE 15, ATARI15, ARABIDOPSIS ARIADNE 15 |
-0.365 |
0.069 |
| 89 |
265297_at |
AT2G14080
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.363 |
0.087 |
| 90 |
246888_at |
AT5G26270
|
unknown |
-0.361 |
0.273 |
| 91 |
249010_at |
AT5G44580
|
unknown |
-0.352 |
-0.018 |
| 92 |
264963_at |
AT1G60600
|
ABC4, ABERRANT CHLOROPLAST DEVELOPMENT 4 |
-0.347 |
-0.240 |
| 93 |
261772_at |
AT1G76240
|
unknown |
-0.342 |
0.097 |
| 94 |
245729_at |
AT1G73490
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.342 |
-0.029 |
| 95 |
258472_at |
AT3G06080
|
TBL10, TRICHOME BIREFRINGENCE-LIKE 10 |
-0.332 |
-0.195 |
| 96 |
249190_at |
AT5G42750
|
BKI1, BRI1 kinase inhibitor 1 |
-0.328 |
-0.444 |
| 97 |
256446_at |
AT3G11110
|
[RING/U-box superfamily protein] |
-0.321 |
-0.210 |
| 98 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
-0.319 |
-0.160 |
| 99 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.299 |
-0.425 |
| 100 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.296 |
0.196 |
| 101 |
259546_at |
AT1G35350
|
[EXS (ERD1/XPR1/SYG1) family protein] |
-0.283 |
0.207 |
| 102 |
251905_at |
AT3G53710
|
AGD6, ARF-GAP domain 6 |
-0.281 |
-0.030 |
| 103 |
256257_at |
AT3G11240
|
ATATE2, ATE2, arginine-tRNA protein transferase 2 |
-0.272 |
0.012 |